Вернуться к статье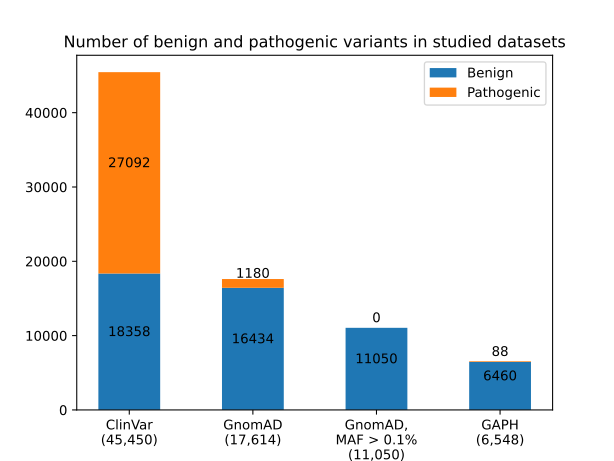
ВЫРАВНИВАНИЕ ПСЕВДОРИДОВ ГОМОЛОГИЧНЫХ ПОСЛЕДОВАТЕЛЬНОСТЕЙ В ОПРЕДЕЛЕНИИ ПОТЕНЦИАЛЬНО ПЕРЕНОСИМЫХ ГЕНОМНЫХ ВАРИАНТОВ
Figure 1 - Distribution of benign and pathogenic variants from ClinVar, GnomAD v3.1.2, GnomAD subset with MAF of variants > 0.1%, and the GAPH-resulted variant set

the total number of benign and pathogenic variants is indicated in parentheses
