ОЦЕНКА ГОМОЛОГИИ HOG1, 14-3-3 И STE11 У ПРЕДСТАВИТЕЛЕЙ СИМБИОТИЧЕСКИХ МИКОРИЗНЫХ ГРИБОВ
ОЦЕНКА ГОМОЛОГИИ HOG1, 14-3-3 И STE11 У ПРЕДСТАВИТЕЛЕЙ СИМБИОТИЧЕСКИХ МИКОРИЗНЫХ ГРИБОВ
Аннотация
Адаптация к засухе древесно-кустарниковых растений связана с видовой спецификой и наличием симбиотических связей с арбускулярной микоризой. В природе отмечается широкое разнообразие арбускулярной микоризы, самыми распространенными таксономическими группами являются: Glomus, Diversispora, Paraglomus, Acaulospora, Entrophospora, Gigaspora, Scutellospora. В запуске механизмов адаптации к стрессу при симбиотической связи «гриб-растение» важную роль играют сигнальные белки Hog1, 14-3-3, Ste11 у представителей арбускулярной микоризы. В связи с этим целью работы стало оценить гомологию аминокислотных последовательностей сигнальных белков Hog1, 14-3-3 и Ste11 у разных симбиотических микоризных грибов. Для исследования были взяты их аминокислотные последовательности из базы данных Protein (NCBI) и при помощи программы BLAST был выполнен поиск гомологов белков в последующем с которыми проводилось множественное выравнивание программой MEGA11 с использованием алгоритма MUSCLE. Филогенетический анализ проводился с помощью программы MEGA11 методом максимального правдоподобия. В результате для белков Hog1, 14-3-3 была выявлена высокая консервативность и высокая вариабельность для Ste11. Также было показано расхождение пары гомологов белка 14-3-3 у Glomeromycotina до отделения подтипа и Ste11 у Glomeraceae после формирования семейства. В итоге мы определили, что Ste11 является перспективным белком интереса для поиска внутривидовых вариаций, ассоциированных с устойчивостью к засухе и прочим абиотическим факторам стресса в системе «гриб-растение».
1. Introduction
Climate change leads to desertification and land degradation, which leads to in biodiversity of woody and shrubby plants decrease . The problem of reducing desertification in southern regions and preventing degradation of agricultural lands stands before Russian science more and more sharply at present times. One of the methods to stop desertification, particularly, sands fixation is planting of several woody and shrubby plants, such as Calligonum aphyllum (Pall.) Gürke, Tamarix ramosissima Ledeb., Haloxylon aphyllum (Minkw.) Iljin, Krascheninnikovia ceratoides L. и Ulmus pumila L. Species features influences crucially on plants adaptation to drought, common in territories with arid climate, but important, sometimes, a key factor is symbiotic mycorrizal fingi presense, which provide nutrition and protection from unfavoravle factors, favor addditional protective mechanisms formation and, so, increasing plant sustainability to stress factors . Mycorrizal arbuscular fungi in C. aphyllum, T. ramosissima, H. aphyllum, K. ceratoides and U. pumila are not yet studied , so their particular role in drought adaptation processes are not yet described. However, we can assume, that in the arid climate those genie ill be amongst mycorrizal species: Glomus, Diversispora, Paraglomus, Acaulospora, Entrophospora, Gigaspora, Scutellospora or other genie of symbiotic fungi. Now there are evidences of arbuscular mycorriza positive influence on plant’s drought tolerance .
Drought tolerance increase was shown for plants Glycine max L., but species diversity of mycorrizal species was not studied in that research . Ipomoea batatas (L.) Lam endured drought easier with inoculation of fungi genie Glomus sp.and Acaulospora sp. . Majority of such resources were carried out on agricultural plants , but endomycorrizal fungi species diversity forming symbiosis with desert and semi-desert plants is not studied alike protective mechanisms formation in plants forming a symbiosis.
Proteins Hog1, 14-3-3, Ste11 are playing important role in drought adaptation mechanisms launching in arbuscular mycorriza fungi in symbiotic bond “fungus-plant”. Hog1 and Ste11 are part of kinase cascade inhibiting large amount of genes, associated with drought tolerance , like genes of aquaporins, regulatory proteins 14-3-3. 14-3-3 is a large grope of proteins, genomes of many organisms contain several homologues of genes of this group. These genes are present in genomes of fungi, plants and animals. In fungi, these genes take part in many processes like neutral thregalase Nth1 activation and change in expression of 220 genes in Saccharomyces cerevisiae genome . Ste 11 is a MAPKKK kinase, signal carrier, participant of several signal pathways. Its involvement in fungi pheromones response , plant infection by pathogenic fungus and drought tolerance was discovered. These proteins are of interest in drought tolerance mechanisms formation research and possibilities of its improvement for plant and fungus system.
Regarding this, the goal of our research was to estimate amino-acid sequence homology of signal proteins Hog1, 14-3-3 and Ste11 amongst different mycorrizal fungi species for further study of symbiotic bond “fungus-plant” formation features in conditions of drought.
2. Research methods and principles
Signal proteins Hog1, 14-3-3, Ste11 presented among diverse species of arbuscular mycorriza were selected for research. Amino-acid sequences for alignment building were taken from Protein (NCBI) database and search for homologous of Hog1, 14-3-3 и Ste11 was carried out using program BLASAT. Protein sequences of Rhizophaus irregular, which genes are annotated and localised in chromosome, were selected as initial query. Homologous annotated as Hog1 were selected using instrument ProteinBLAST , in case of absence of annotated homologous – closest uncharacterized homology. Those species were selected to build an alignment:
1) Rhizophaus irregularis (XP_025185210.1);
2) Rhizophaus diaphanus (RGB42528.1);
3) Rhizophaus clarus (GBB87021.1);
4) Funneliformis geosporum (CAI2181536.1);
5) Glomus cerebriforme (RIA92778.1);
6) Geosyphon pyriformis (KAG9291825.1);
7) Ambispora gerdemannii (CAG8436886.1);
8) Diversispora epigaea (RHZ50009.1);
9) Acaulospora morrowiae (CAG8446886.1);
10) Racocetra persica (CAG8495386.1);
11) Cetraspora pellucida (CAG8501949.1);
12) Dentiscutata erythropus (CAG8739878.1);
13) Umbelopsis sp. PIM 123. (KAH8549606.1).
Proteins 14-3-3 and Ste11 were selected according to the same principle, but with slight changes in the species of the presented proteins, which are associated with the presence or absence of characterized amino acid sequences in the NCBI Protein databases
. 14-3-3 proteins of the species were taken:1) Rhizophagus irregularis DAOM (XP 025173882.1);
2) Rhizophaus irregularis (CAJ16742.1, PKY38380.1);
3) Glomus cerebriforme (RIA97653.1);
4) Gigaspora rosea (RIB16586.1);
5) Gigaspora margarita (KAF0484528.1);
6) Gigaspora rosea (RIB2229.1);
7) Glomus cerebriforme (RIA79980.1);
8) Gigaspora margarita (KAF0420307.1);
9) Funneliformis caledonium (CAG8442663.1);
10) Funneliformis geosporum (CAI2173046.1);
11) Acaulospora morrowiae (CAG8439107.1);
12) Cetraspora pellucida (CAG8612119.1);
13) Diversispora Epigaea (RHZ47341.1);
14) Ambispora gerdemannii (CAG8474580.1);
15) Geosiphon pyriformis (KAG9289207.1);
16) Dentiscutata erythropus (CAG8685133.1);
17) Racocetra persica (CAG8610165.1);
18) Umbelopsis sp PMI 123 (KAH8548527.1).
To search for the closest Ste11 homologues, the following amino acid sequences were taken from mycorrhizal symbiotic fungi:
1) Rhizophagus irregularis DAOM (XP 025186107.1);
2) Rhizophaus irregularis (XP 025172837.1);
3) Rhizophagus diaphanus (RGB34169.1, RGB38268.1);
4) Rhizophagus clarus (GES82803, GBB91729.1);
5) Funneliformis caledonium (CAG8474800.1, CAG8456822.1);
6) Funneliformis geosporum (CAI2186005.1);
7) Acaulospora morrowiae (CAG8664532.1, CAG8489781.1);
8) Glomus cerebriforme (RIA89482.1);
9) Dentiscutata erythropus (CAG8619186.1);
10) Gigaspora rosea (RIB01303.1);
11) Gigaspora margarita (CAG8472639.1);
12) Geosiphon pyriformis (KAG9304061.1);
13) Ambispora leptoticha (CAG8599465.1);
14) Diversispora Epigaea (RHZ44604.1);
15) Racocetra fulgida (CAG8484893.1);
16) Umbelopsis sp PMI 123 (CAG8583455.1, KAH8553767.1).
To identify differences between signaling proteins, bioinformatic analysis was performed. It included multiple alignment of the amino acid sequences of the studied proteins using the Mega 11 program (MEGA, Japan) according to the MUSCLE algorithm with default settings: penalty for gap opening -2.90, penalty for gap expansion 0, hydrophobicity multiplier 1.20. Alignment filtering was done manually with minimal changes: only the terminal unaligned regions were removed. Also, using MEGA 11, the most suitable model of amino acid substitutions was selected: LG + G for Hog 1 and 14-3-3, and JTT + G + F for Ste 11. Based on the amino acid sequences, a phylogenetic tree was constructed using the maximum likelihood method with bootstrap estimation -support, in the MEGA11 program (MEGA, Japan).
3. Main results and Discussion

Figure 1 - N-terminus of Hog1 alignment

Figure 2 - C-terminal variable regions of Hog1 proteins
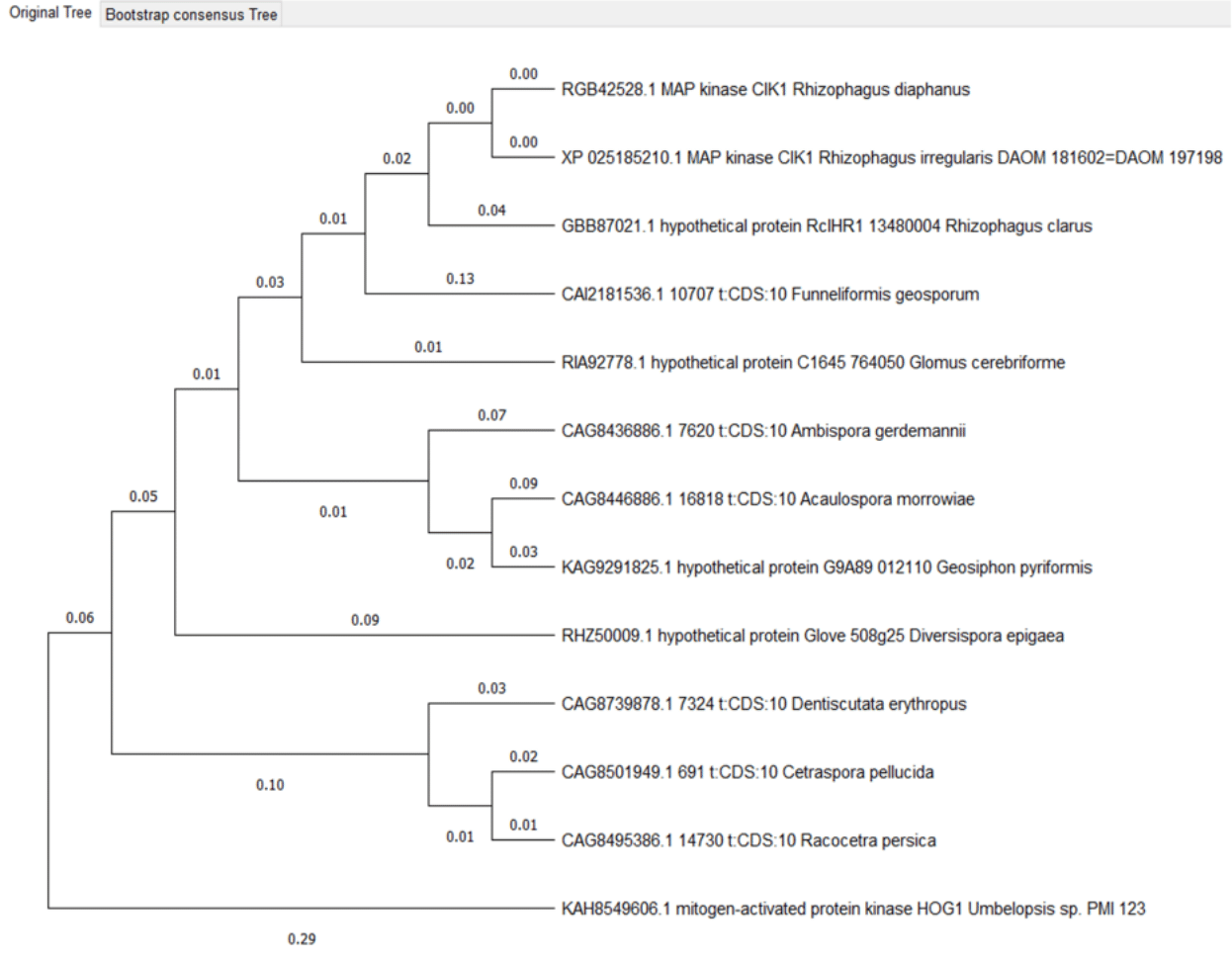
Figure 3 - Hog 1 proteins phylogenetic tree
The clade consisting of the proteins Geosyphon pyriformis, Ambispora gerdemannii and Acaulospora morrowiae differs from the phylogenetic tree of organisms, Ambispora and Geosyphon belong to the order Archaeosporales, and Acaulospora belongs to the order Diversisporales and the family Acaulosporaceae.
14-3-3 protein is also conserved, featured by a small variable region at the N-terminus, as well as a longer sequence in Cetraspora pellucida, far beyond the general conserved part of the protein at the N-terminus. (Fig. 4)
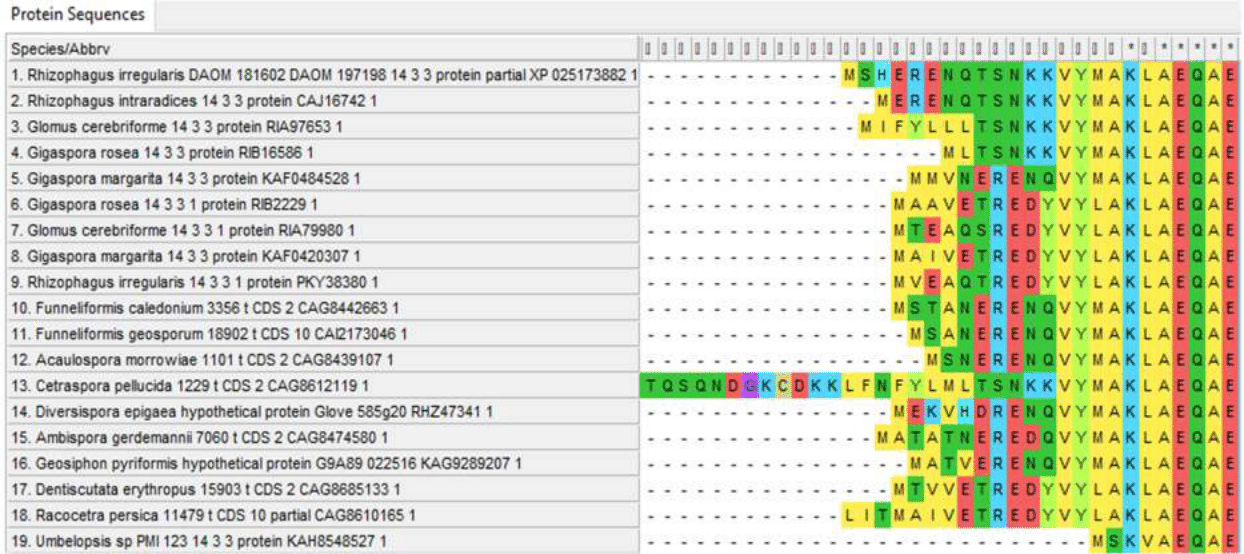
Figure 4 - N-terminal regions of 14-3-3 proteins
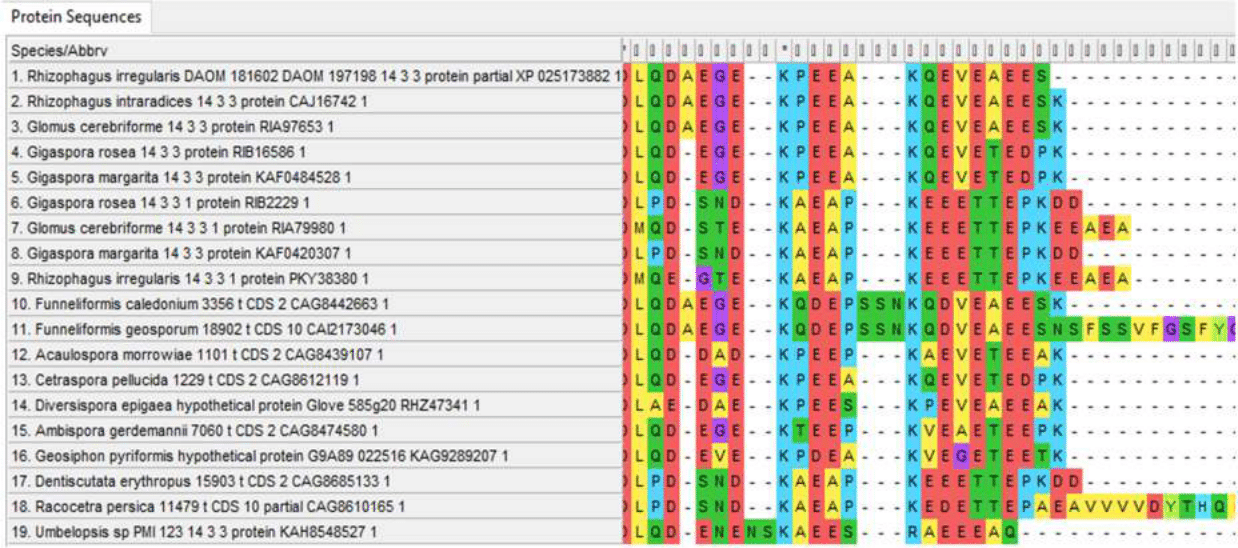
Figure 5 - Variable C-terminal regions of 14-3-3 protein

Figure 6 - 14-3-3 proteins phylogenetic tree
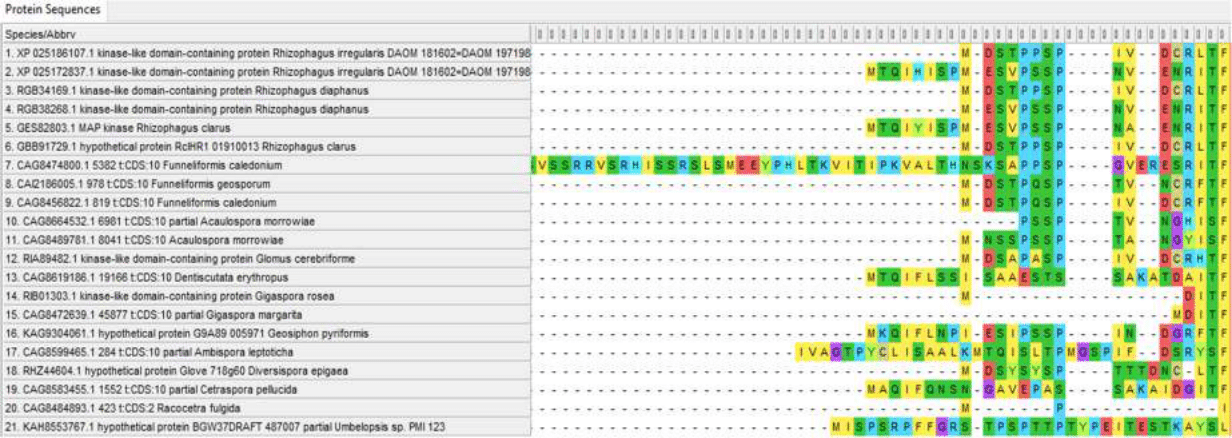
Figure 7 - Variable N-terminus of Ste11 proteins
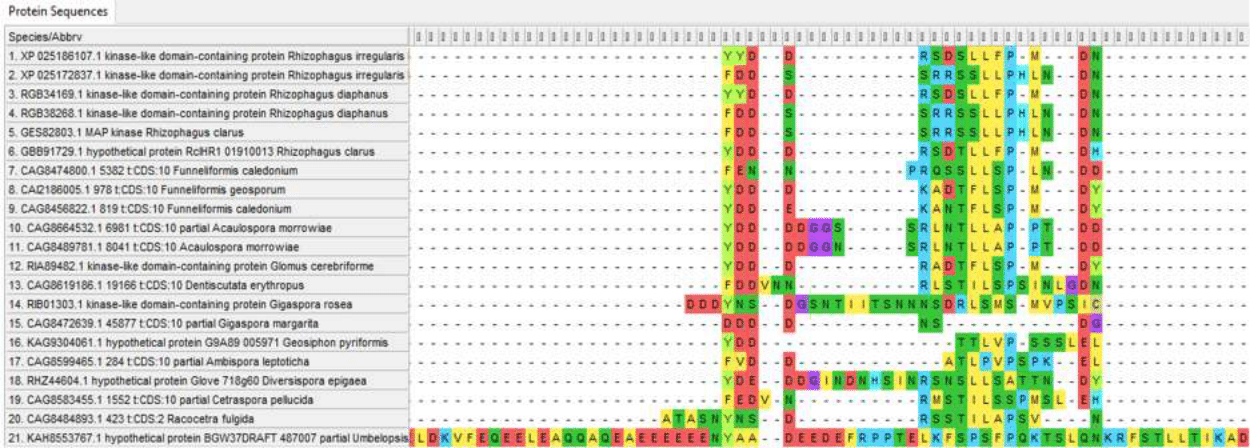
Figure 8 - Insertions and variable regions in the internal part of Ste11 proteins amino-acid sequence

Figure 9 - Ste11 proteins phylogenetic tree
However, variability in the sequences of these proteins is present not only at the ends of the chain, which is typical for most proteins, but also in the central part. And although at the moment there is no data on the variability of the amino acid sequences of these proteins within species and the differences between different types of endomycorrhizal fungi in relation to the drought resistance factor, such data can be obtained in the framework of further studies, which leaves hope for linking variations in these proteins with the resistance of the “fungus-plan” pair to drought and its mechanisms.
Promoter regions of the genes of the studied proteins also require attention. Since the proteins themselves are highly conserved, markers for selecting symbiotic fungi as inocula should be sought in the promoter regions of their genes, attention to upstream and downstream regulators should be paid as well.
The Ste11 protein is much more variable; the distance to other proteins can reach 0.80988 (Umbelopsis and Diversispora epigaea); within Glomeromycotina, distances often exceed 0.4. This variability of the protein gives hope for detecting variations in its sequence associated with drought resistance of the “fungus-plant” system, which makes it a priority object for further research.
4. Conclusion
Alignments and phylogenetic trees were constructed for the Hog1, 14-3-3 and Ste11 proteins, which showed high conservativity of the Hog1 14-3-3 proteins and variability of the Ste11 protein. The divergence of the homologue pair 14-3-3 in Glomeromycotina before subphylum separation and Ste11 in Glomeraceae after the formation of the family was shown. Ste11 has been identified as a promising protein of interest for the search for intraspecific variations associated with resistance to drought and other abiotic stress factors.
