IN SILICO DESIGN OF PRIMERS FOR PROFILING SPARE SOY PROTEINS
IN SILICO DESIGN OF PRIMERS FOR PROFILING SPARE SOY PROTEINS
Abstract
Soybean is an important agricultural product that has become widespread and popular due to its high nutritional qualities, namely protein content. One of the problems faced by commodity producers is a large number of so–called anti-nutritional proteins, trypsin and chymotrypsin inhibitors, due to the high concentration of which additional heat treatment is required before eating or for animal feed. Useful proteins include, in some cases, up to 70% of the total protein content of glycinins and b-conglycinins, which are a group of storage proteins. The creation of soybean varieties with an increased content of storing proteins requires a genetic analysis of the breeding material to identify the presence and level of expression of glycinins and b-conglycinins mRNA. This requires modeling of the most specific and not prone to the formation of secondary structures of primers. To model them, transcripts of the loci of interest for glycinins (Gy1-Gy5, Gy7) and β-conglycinins (CG-1, CG-4, CG-3) were found in the PlantsEnsemble database. The next step on the Unipro Ugene platform using the Primer3 toolkit was to select many pairs of primers for each of the loci, after which the pairs of primers were tested for specificity using Primer Blast from NCBI and for the formation of secondary structures (hairpins, homo- and heterodimers). As a result of the work done, pairs of primers were obtained that can be used to detect and determine the expression level of the genes of interest to us.
1. Introduction
Soy is a high-protein product in demand, used in the manufacture of food for humans and feed for farm animals. Soy products are of particular value due to their high protein content, despite the presence of anti-nutritional elements, trypsin and urease inhibitors among them. Of particular interest among the protein diversity are the so-called storage proteins, concentrated in soybean seeds and playing an important role in seed germination. Glycine proteins make an important contribution to the nutritional value of soy. Soybean seeds differ from other legumes in a particularly high protein content, accounting for an average of 40% of the total dry matter and, therefore, are an important source of vegetable protein both in nutrition and in industry. Two main spare proteins, conglycinin and glycinin, corresponding to 7S and 11S globulins, make up about 70% of the total protein of soybean seeds , .
Glycine (11S) and b-conglycine (7S) are two representatives of the class of storage proteins and account for approximately 70% of the total protein in ordinary soybean seed. Proteins of this family are of great nutritional and economic importance. To date, significant studies of the genes encoding these proteins have been carried out, since their in-depth study may become a new direction in the molecular breeding of soybeans , , .
Glycine is a hexameric protein consisting of six similar subunits, each of which includes an acidic polypeptide chain linked by a disulfide bond to the main polypeptide chain. Several genes have been discovered that encode these subunits. The literature describes highly homologous forms of glycine, designated as Gy1, Gy2 and Gy3 (group I). Later, the presence of two additional glycine genes was discovered: Gy4 and Gy5 (group II). These five genes in the above two groups encode the predominant subunits of glycine hexamers. All genes consist of four exons and three introns, and the sequence identity is about 45% between the two above groups and about 80% within each group. In addition, two additional glycine genes were later identified: one Gy6, which is a pseudogen and is Gy7, a gene with weak expression. b-Conglycinin is a trimeric protein consisting of the α, α’ and β subunits. 15 genes related to these subunits have been identified. They are designated from CG-1 to CG-15. These genes are divided into two large groups, united into several genetic regions. Of these genes, CG-1 encodes the a’ subunit, CG-4 encodes the b subunit, and CG-2 and CG-3 encode a subunit. However, the remaining genes have not been studied to a large extent, and it is unknown to which linkage group they belong, regardless of the functional activity and the encoded subunit (α, α’ and β) , , .
Profiling soybean varieties by the presence of genes, the products of which are various subunits of storage proteins, can help further breeding work aimed at obtaining the most favorable soybean varieties for human nutrition and animal feeding.
Real-time PCR technology can help to cope with this task, which will not only detect the presence of certain genes, but also determine the level of their expression. This requires the selection of the most suitable pairs of primers that do not form homo- and heterodimers, as well as specifically binding to their target sequences of the mRNAs of interest.
The objective of our work was to create mRNA-specific primers of various glycinin and β-conglycinin subunits that could be used for profiling cultivated soybeans.
2. Research methods and principles
The search for the loci of interest took place in the Plants Ensemble database. The obtained transcript data were compared with the data on the presence of the proteins themselves in Uniprot. Further, information about the primary sequence of nucleotides was exported to the Unipro Ugene platform. The selection of primers was carried out using the integrated Primer3 plugin with the following settings: amplicon length 200-500 n.p., primer size from 18-27 n.p. with an average value of 20 n.p., the melting point of primers from 50–60 degrees, with an optimum of 55, GC composition from 40-60%, with a maximum self–complementarity of no more than 4 n.p. using the thermodynamic model SantaLucia 1998. The obtained primer pairs were selected using the OligoAnalyzer web toolkit, with the following parameters: mg 3 mM sodium 50 and oligo 0.04. The selected primers underwent additional specificity testing on the NCBI service using the Primer Blast tool.
3. Results and Discussion
As a result of searching for transcriptome sequences of genes of interest to us in the PlantsEnsemble database, the loci necessary for modeling primers were found: Gy1 (KRH67385), Gy2 (KRH67383), Gy3 (KRG95667), Gy4 (KRH32185), Gy5 (KRH19559), Gy7 (KRG95666), Cg-1(KRH35498), Cg-3(KRG91330), Cg-4(KRG91303). The next step on the Unipro Ugene platform, using the Primer3 plugin, was to compile a list of primer pairs for each transcript. The obtained pairs of primers underwent a number of additional checks for the formation of undesirable structures, such as hairpins, homo- and heterodimers. Information about the most successful primers and the characteristics of secondary structures are shown in table 1.
Table 1 - The most specific primers
Locus | Primer | Product length (bp) | Dimers (kcal/mol;bp) | Hairpin (kcal.mole; oC) |
Gy1 | F: GCAAGAGCAAGGAGGTCATC R: TTGCACTGTGGCTTCTCATC | 285 | Homo-dimer F: -3,14;2 R: -7,05;4 Hetero-Dimer -7,04;4 | F: -1.01;42,3 R: -0.82;36.1 |
Gy2 | F: AGCCAGAAAGGAAAGCAACA R: GGCACTGAGTTTGAGAAGCC | 444 | Homo-dimer F: -3,14;2 R: -6,21;3 Hetero-Dimer -6,21;3 | F: -1.51;52,3 R: -0,94;39,2 |
Gy3 | F: CGAAGCCACCTTACACCATT R: GGCTTGTTGTTAGGGTTCCA | 392 | Homo-dimer F: -3,61;2 R: -3,14;2 Hetero-Dimer -9.75;5 | F: -0,12;27 R: 0,23;21,2 |
Gy4 | F: GCTTGGTCGACTAGGACGAG R: CTCCAGACGACGAAAGGAAG | 221 | Homo-dimer F:-9,45;6 R: -4,64;3 Hetero-Dimer -6,53;4 | F: -0,91;37,5 R: -0,27;29,9 |
Gy5 | F: CAACAGCAGCAGCAGAAGAG R: CACGTGGTTCTTGTTGTTCG | 403 | Homo-dimer F: -3,14;2 R: -10,2;6 Hetero-Dimer -7,19;5 | F: -2,23;57.4 R: -0,1;26,3 |
Gy7 | F: CACCCTTGGAACACGAAGTT R: GTTGAATATGTCGGCGTGTG | 392 | Homo-dimer F: -3,61;2 R: -3,91;4 Hetero-Dimer -8.25;5 | F: -0,98 R: 0,45;18 |
Cg-1 | F: TGCTGGGACTTGTTTTCCTGGCT R: ACTCCCTCTTGCTGCCTCTGCT | 1096 | Homo-dimer F: -4,64;3 R: -3,14;2 Hetero-Dimer -7,71;4 | F: -3,12;47,6 R: -1,09;40,3 |
Cg-3 | F: GCTGGGAGTTGTTTTCCTGGCA R: TTGGTGTGGGCGTGGGTGTT | 430 | Homo-dimer F: -4,46;3 R: -3,61;2 Hetero-Dimer -3,14;2 | F: -3;48 R: -1,12;5 |
Cg-4 | F: ACTCCTACAACCTTCACCCTGGCG R: ACTCCCTCTTGCTGCCTCTGCT | 286 | Homo-dimer F: -3,61;2 R: -3,14;2 Hetero-Dimer -6,21;3 | F: -0,09;26,3 R: -1,09;40,3 |
The formation of undesirable structures can affect the final picture obtained as a result of the study by the appearance of fragments with different molecular weights, which makes it difficult or impossible to detect the target product . Based on this, it is worth paying not a little attention to the possibility of the formation of hairpins, homo- and heterodimers, which requires a number of additional studies on services such as Oligoanalizer (idtDNA) and RNAfold (fig. 1).
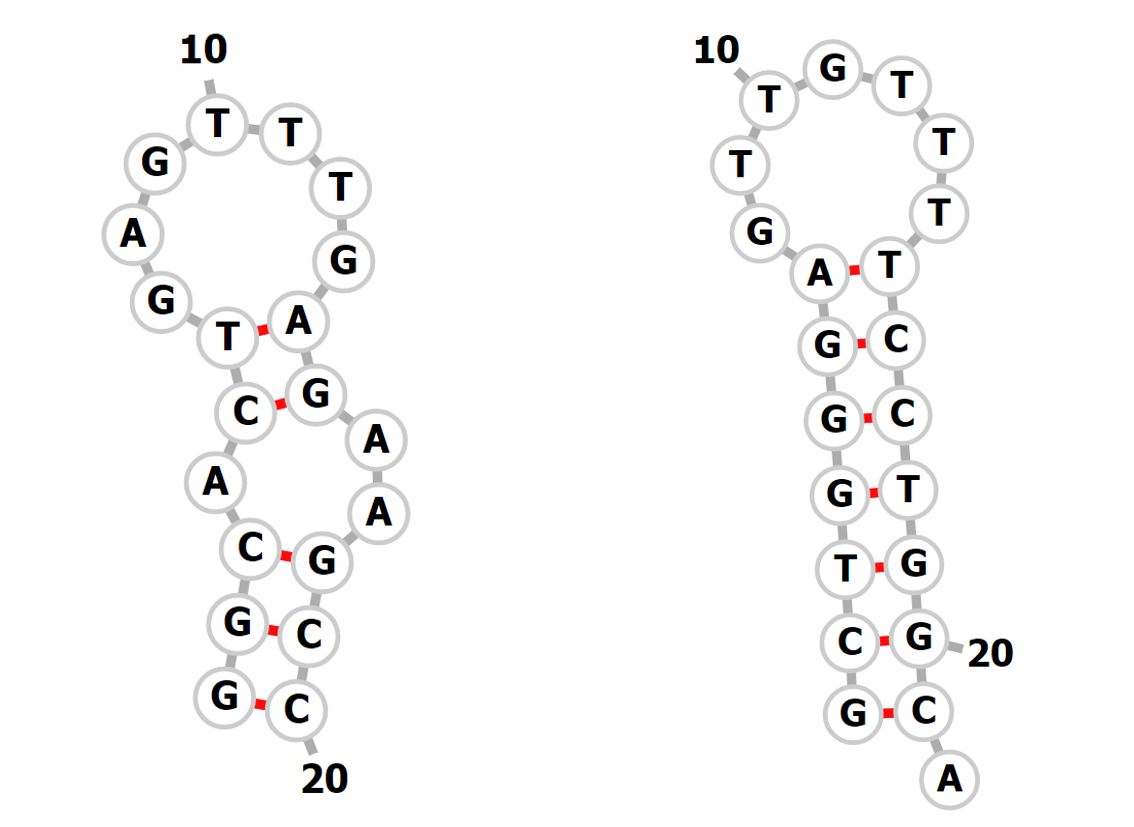
Figure 1 - Example of unwanted secondary structures
It was found that some primers form secondary structures, but the binding energy of these structures is too small, which allows us to conclude that their occurrence is unlikely. Figure 1, as an example, shows the hairpins that can form in primers for glycine (Gy2) and conglycinin (Cg-3) loci. The presented structures have a low value of the binding energy modulus, namely -0.94 for G5 and -3; 48 for Cg-3, which indicates a low probability of the appearance of these structures.
One of the most important parameters for a pair of primers is its specificity, since the detection of non-target targets can lead to a false positive result . To avoid this, the obtained primer pairs were analyzed using the NCBI Primer Blast web tool. The conducted check showed high specificity of the selected pairs of primers.
4. Conclusion
As a result of the work done, pairs of primers flanking the loci of glycinin (G1, G2, G3, G4, G5, G7) and conglycinin (Cg-1, Cg-3, Cg-4) were obtained. Primers meet the requirements of specificity, and also, for the most part, are not prone to the formation of stable secondary structures under the conditions of PCR.
