ОЦЕНКА СТРУКТУРНОЙ ГОМОЛОГИИ МЕЖДУ ФУНКЦИОНАЛЬНО СХОДНЫМИ БЕЛКАМИ ВЫСОКОСПЕЦИАЛИЗИРОВАННЫХ ТКАНЕЙ НА ПРИМЕРЕ ПРОТЕОГЛИКАНОВ И БЕЛКОВ СУРФАКТАНТА
DOI: https://www.doi.org/10.18454/jbg.2023.3.19.001
Krylov P.A.1 *, Tretyakova A.V.2, Gerasimova E.O.3, Fetisova E.I.4, Novochadov V.V.5
1-5 Volgograd State University, Volgograd, Russia;
1 Federal Scientific Center for Ecology, Integrated Land Reclamation and Protective Aforestation of the Russian Academy of Sciences, Volgograd, Russia
*Corresponding author (krylov.pavel[at]volsu.ru)
Received: 02.11.2022; Accepted: 02.12.2022; Published: 27.02.2023
EVALUATION OF STRUCTURAL HOMOLOGY BETWEEN FUNCTIONALLY SIMILAR PROTEINS OF HIGHLY SPECIALIZED TISSUES ON THE EXAMPLE OF PROTEOGLYCANS AND SURFACTANT PROTEINS
Research article
Abstract
Proteins synthesized by cells of various tissues may have common functions, but at the same time have a heterologous structure. Previously, it was experimentally confirmed that lung suffractant proteins can be synthesized by chondrocytes of articular cartilage. Articular cartilage proteoglycans, such as lubricin (PRG4) and aggrecan (ACAN), have partially similar functions to surfactant proteins (SP-A, SP-B, SP-C, and SP-D). The main goal of the work was to evaluate of structural homology between these proteins. The Ugene program was the source of the of multiple and paired alignment results using the ClustalW and Smith–Waterman algorithm, and the MEGA11 program provided phylogenetic analysis. A common domain of the Lectin C-type family with a high degree of similarity to the domains of EGF-like proteoglycans (ACAN) and SMB 1 (PRG4) was found in ALAN, SP and SP-D proteins. The Sasposin domains of the SP-B protein had the greatest similarity with the PRG4 and ACAN domains over 57%. The BRICHOS domain of the SP-C protein had similarities with the SMB1, SMB2 (PRG4) and EGF-like (ACAN) domains. The mucin domain in the PRG4 structure was not detected. The phylogenetic analysis revealed about stidied proteins to have low level of evolution homology, as evidenced by bootstrap support.
Keywords: multiple alignment, ClustalW, PRG4, ACAN, SP-A, SP-B, SP-C, SP-D, mucin, articular cartilage.
Крылов П.А.1 *, Третьякова А.В.2, Герасимова Е.О.3, Фетисова Е.И.4, Новочадов В.В.5
1-5 Волгоградский государственный университет, Волгоград, Россия;
1 Федеральный научный центр агроэкологии, комплексных мелиораций и защитного лесоразведения Российской академии наук, Волгоград, Россия
* Корреспондирующий автор (krylov.pavel[at]volsu.ru)
Получена: 02.11.2022; Доработана: 02.12.2022; Опубликована: 27.02.2023
ОЦЕНКА СТРУКТУРНОЙ ГОМОЛОГИИ МЕЖДУ ФУНКЦИОНАЛЬНО СХОДНЫМИ БЕЛКАМИ ВЫСОКОСПЕЦИАЛИЗИРОВАННЫХ ТКАНЕЙ НА ПРИМЕРЕ ПРОТЕОГЛИКАНОВ И БЕЛКОВ СУРФАКТАНТА
Научная статья
Аннотация
Белки, синтезируемые клетками различных тканей, могут обладать общими функциями, но при этом иметь гетерологичную структуру. Ранее было экспериментально подтверждено, что белки суфрактанта легких могут синтезироваться хондроцитами суставного хряща. Протеогликаны суставного хряща, такие как лубрицин (PRG4) и аггрекан (ACAN), имеют частично сходные функции с белками сурфактанта (SP-A, SP-B, SP-C и SP-D). Основной целью работы стал поиск структурной гомологии между этими белками. Программа Ugene являлась источником результатов множественного и парного выравнивания при использовании алгоритмом ClustalW и Smith–Waterman, а программа MEGA11 обеспечивала проведение филогенетического анализа. В белках ACAN, SP-A и SP-D был обнаружен общий домен семейства Lectin C-type с высокой степенью схожести с доменами протеогликанов EGF-like (ACAN) и SMB 1 (PRG4). Домены Sasposin белка SP-B имели наибольшее сходство с доменами PRG4 и ACAN свыше 57%. Домен BRICHOS белка SP-C имел сходство с доменами SMB1, SMB2 (PRG4) и EGF-like (ACAN). Муциновый домен в структуре PRG4 не был обнаружен. В результате филогенетического анализа обнаружилось, что изученные белки демонстрируют низкий уровень эволюционной гомологии, о чём свидетельствуют результаты бутстрапподдержки.
Ключевые слова: множественное выравнивание, ClustalW, PRG4, ACAN, SP-A, SP-B, SP-C, SP-D, муцин, суставной хрящ.
1. Introduction
The fact that a wide variety of functions prompt through a very limited number of solutions in polymer structures is a wellknown law of the molecular organization in living organisms. The inevitability of mutations, which are the leading, not the only, mechanism of molecular evolution, leads to the fact that proteins with a similar function, having both homologous and heterologous structures, can exist in various tissues of the body. Finding out the causes and possible mechanisms of the appearance of the first and second variants of proteins with a similar function can clarify a number of features providing the tissues specialization in a multicellular organism and specify the molecular evolution mechanisms of some protein groups in connection with its functions.
The present study focuses on the structural and functional homology between the proteoglycans of the superficial and intermediate zones of articular cartilage [1], as well as four pulmonary surfactant proteins [2].
The interest in these proteins is due to fact that surfactant proteins can be applied as premixes to existing drugs used for intra-articular injections in osteoarthritis (OA). It is in concordance to common strategy of OA treatment moving away from the use of invasive to minimally invasive methods [3]. Viscosapplementary therapy or intra-articular injections are minimally invasive methods of treating OA, since they aimed at simplifing drug delivery, providing anti-inflammatory effect, and improvement of viscoelastic properties of both synovial fluid and articular cartilage surface [4]. Earlier, we showed in pilot studies that the addition of surfactant proteins to the synovial fluid increases its tribological characteristics in vitro [5], it reduces the articular cartilage damage in experimental OA in vivo [6].
Disturbances of the synthesis for each studied protein can contribute to the development of diseases. For example, a decrease in the number of proteoglycans in articular cartilage is an integral part of the development of OA [7], [8], which affects approximately 15 millions people in Russia and above 300 millions people in world [9], [10]. Disturbances of surfactant protein synthesis are associated with a fairly wide spectrum of pulmonary diseases [11]. Published experimental data show that chondrocytes synthesize surfactant proteins, both in normal and in the development of OA [8]. These facts are the basis to search for a possible structural homology between the described proteins, taking into account the similarity of their functions.
Lubricin (PRG4) [13], [14] and surfactant proteins B and C (SP-B and SP-C) [15], [16], [17] participate in the film formation on the articular and alveolar surfaces to ensure a reduction in the sliding friction coefficient, surface tension, and other tribological characteristics. Other experimental data demonstrate that PRG4 has mucin-like domains/repeats in its structure [14], [18], which give it high lubricative properties. Aggrecan (ACAN) [15], [19] and surfactant proteins A and D (SP-A and SP-D) [11], [20] perform structure-forming function and some tissue-specific functions.
Based on the above, the purpose of this study is to evaluate the structural homology between proteoglycans and surfactant proteins using current bioinformatics tools.
2. Materials and methods
2.1. Data sources for bioinformatics analysis
Open-access databases UniProtKB [27], NCBI Protein [28], EMBL-EBI [29] and Ensembl [30] were a source of information about the amino acid sequences of the studied proteins and their domains.
To analyze the differences between Human and mammalian proteins, we took the complete amino acid sequences of the studied proteins of ten biological species. There are some criteria to choose these species. Baboon (Papioanubis) was choosen as close organism to humans (Homo sapiens). The bull (Bos taurus), sheep (Ovis aries), and pig (Sus scrofa) acted at animals widely used in agriculture. Pacific walrus (Odobenus rosmarusdivergens) and blue whale (Balaenoptera musculus) were primers of animals, mainly leading an aquatic lifestyle with loads on articular cartilage, different from animals living on land. Three rodents used in research practice as gray rat (Rattus norvegicus), Syrian hamster (Mesocricetus auratus), and domestic mouse (Mus musculus) completed our species selection (Table 1).
Table 1 – Entry identifiers of the studied organisms
|
Organisms |
|
Proteins and their identifiers |
|
|
||
|
ACAN |
PRG4 |
SP-A |
SP-B |
SP-C |
SP-D |
|
|
Homo sapiens |
P16112 |
Q92954 |
Q8IWL2-2 |
P07988 |
P11686 |
P35247 |
|
Papioanubis |
A0A096NN51 |
A0A2I3N8Z7 |
A0A2I3LG07 |
A0A096NZV5 |
A0A096N8K2 |
A0A096P4G4 |
|
Bos taurus |
P13608 |
A0A3Q1LZ67 |
Q6RXL1 |
P15781 |
P15783 |
P35246 |
|
Ovis aries |
W5PNK3 |
W5P880 |
Q9TT06 |
A0A6P7DVV5 |
Q9N276 |
A0A6P7DGQ9 |
|
Sus scrofa |
A0A287B863 |
XP_020919476.1 |
P49874 |
A0A4X1W7A7 |
A0A8D0WYX0 |
Q9N1X4 |
|
Odobenus rosmarus divergens |
A0A2U3ZDN8 |
A0A2U3WBI2 |
A0A2U3W5I6 |
A0A2U3X0N9 |
A0A2U3W2Q0 |
A0A2U3W5L7 |
|
Balaenoptera musculus |
A0A8B8WQ24 |
A0A8B8X1A4 |
A0A8B8VFG3 |
A0A8B8ZAA8 |
A0A8B8XPP8 |
A0A8B8VLH4 |
|
Rattus norvegicus |
P07897 |
F1LRA5 |
P08427 |
P22355 |
P11685 |
P35248 |
|
Mesocricetus auratus |
A0A1U8CFJ0 |
A0A3Q0CW00 |
A0A3Q0D8R7 |
A0A1U7QKZ9 |
A0A3Q0CRM2 |
A0A1U8CAZ6 |
|
Mus musculus |
Q61282 |
Q9JM99 |
P35242 |
P50405 |
P21841 |
P50404 |
For a separate study, we took only amino acid sequences of human domains, since the comparison of the domain structure between different mammalians is limited by the completeness of structural annotations for some of the studied proteins and their domains. The selected list of domains included SMB1 and SMB2 for PRG4, EGF-like and Lectin C-type for ACAN, Collagenlike and Lectin C-type for SP-A and SP-D, Sasposin A, Sasposin B1, Sasposin B2, Sasposin B3 for SP-B, and BRICHOS for SP-C. The ACAN Ig-like V-type and SUSHI protein domains were excluded, since they are associated with the performance of immune defense functions.
To evaluate the homologous sites between mucis and PRG4, we took amino acid sequences of mucins (1-22) from the UniProtKB and NCBI Protein databases [28], as well as a repeat |EPAPTTPK| [18] with various variations.
2.2. Bioinformatics methods
The analysis included multiple alignment of amino acid sequences of the studied proteins using the Ugene program (UNIPRO, Russia), the ClustalW algorithm (BLOSUM62 matrix) and paired alignment of domains (the Smith–Waterman algorithm, PAM250 matrix). The similarity of proteins and domains was evaluated in percentage and score. To search for homologous sites of mucin domains MUC (1-22) in the PRG4 protein we performed analisys using the BlastTP program (NCBI, USA) with Max target sequences. Next, we evaluated the presence of common domains in the Conserved Domains program (NCBI, USA) [31].
Phylogenetic trees were constructed based on the maximum likelihood method using the MEGA11 program (MEGA, Japan) with various replacement models. The program MEGA11 using its option Find Best DNA/Protein Models» allowed to to find the best model based on lowest BIC value (Bayesian Information Criterion) as selected feature. To construct a phylogram of the studied human proteins we used the WAG model (Whelan And Goldman model). The alignment of amino acid sequences was performed using the MUSCLE algorithm in the MEGA11 program.
The site coverage less than 95% was the reason for excluding of all positions from analysis, i.e., alignment gaps less than 5%, missing data, and ambiguous bases were allowed at any position as partial deletion options. The scale corresponded to 50 substitutions per 1000 amino acid residues for a phylogram of all human proteins. Bootstrap support used to evaluate the topology, we considered reliable values of branch divergence over 70.
3. Results and discussion
3.1. Comparative analysis of the complete amino acid sequences of the studied human and mammalian proteins
The multiple alignment of proteoglycans and surfactant proteins allowed revealed the degree of homology between human and mammalian proteins (Table 2).
Homo sapien and Papio Anubis have the greatest protein similarity, ranging from 85% to 95%.
Table 2 – Similarity of amino acid sequences of studied proteoglycans and surfactant proteins according to the results of multiple alignment
|
Organisms |
|
|
Proteins |
|
|
|
|
|||||
|
ACAN |
PRG4 |
SP-A |
SP-B |
SP-C |
|
SP-D |
|
|||||
|
% |
score |
% |
score |
% |
score |
% |
score |
% |
score |
% |
score |
|
|
Papioanubis |
87 |
3154 |
85 |
1761 |
85 |
240 |
91 |
424 |
93 |
260 |
95 |
394 |
|
Bos taurus |
78 |
2825 |
57 |
1200 |
79 |
223 |
76 |
351 |
86 |
239 |
76 |
315 |
|
Ovis aries |
76 |
2732 |
69 |
1444 |
78 |
222 |
77 |
357 |
87 |
242 |
76 |
315 |
|
Sus scrofa |
79 |
2875 |
50 |
1065 |
77 |
217 |
73 |
341 |
63 |
176 |
79 |
328 |
|
Odobenus rosmarusdivergens |
77 |
2779 |
67 |
1408 |
65 |
185 |
75 |
346 |
82 |
229 |
79 |
328 |
|
Balaenoptera musculus |
76 |
2762 |
61 |
1274 |
77 |
219 |
75 |
349 |
73 |
203 |
77 |
321 |
|
Rattus norvegicus |
74 |
2678 |
69 |
1439 |
75 |
211 |
74 |
343 |
85 |
236 |
77 |
321 |
|
Mesocricetus auratus |
75 |
2715 |
66 |
1375 |
76 |
214 |
74 |
343 |
85 |
236 |
71 |
296 |
|
Mus musculus |
74 |
2689 |
67 |
1403 |
75 |
211 |
75 |
348 |
84 |
235 |
77 |
320 |
Besides Papioanubis, the amino acid sequence similarity of the ACAN protein between humans and other mammals ranged from 74% to 79% and was maximal for Sus scrofa and minimal one for protein Rattus norvegicus and Mus musculus. The PRG4 similarity analysis yielded lower for all proteins investigated ranging from 50% in Sus scrofa to 69% in Ovis aries and Rattus norvegicus.
The SP-A, SP-B, and SP-D showed akin similarities, ranging near 70%, except for SP-A in Odobenus rosmarus divergens, where it was at 65%. Human SP-C exhibited the highest similarity amino acid sequence with the SP-C sequences Bos Taurus, Ovis aries as well as rodents, where the similarity exceeded 80%. The lowest similarity value was recorded between the SP-C sequences of Human and Balaenoptera musculus.
An interesting finding is the fact that surfactant proteins from farm animals, which are widely used in the pharmaceutical industry and in a number of biotechnological industries [21], have a high degree of homology with human proteins. The high similarity of the SP-C protein sequences in all studied mammals and humans may be due to the fact that this protein appeared relatively late in evolution and is rather conservative in structure [17].
Surfactant proteins from farm animals had been a high degree of homology with human proteins they confirmed to widely used in the pharmaceutical industry and in a number of biotechnological industries [21] The high degree structural homology SP-C between mammals and humans may be due to the fact that this protein appeared relatively late in evolution and is rather conservative in structure [17].
Since proteoglycans and surfactant proteins have different lengths of amino acid sequences: ACAN (3618), PRG4 (2019), SP-A (283), SP-B (464), SP-C (279), SP-D (415), the obtained results of multiple alignment and the comments made on their basis are preliminary. To verify and refine these findings, we carried out the multiple alignment of the studied human proteins at the domain level.
3.2. Structural analysis of the domains of the studied human proteins
The partial pair alignment of proteoglycans and surfactant proteins determined the level of homology and differences between the domains of the studied human proteins were revealed (Table 3, fig. 1, fig. 2).
As can be seen from the presented data, SMB1 and SMB2 domains of the PRG4 had over 40% similarity with the Lectin Ctype domains of the SP-A and the SP-D. The EGF-like domain of the ACAN had the highest similarity value to the Lectin Ctype of the SP-A. At the same time, the homology of the Lectin C-type domain between the SP-A and ACAN was minimal. Homology over 50% was found between the Lectin C-type domain of the ACAN and the Collagen-like domain of SP-A.
The results obtained by pair alignment of the domains of the PRG 4 and the EGF-like domain of the ACAN with the domains of the SP-B protein showed the highest similarity, which ranged from 57% to 62%. At the same time, the similarity of the Lectin C-type domain of the ACAN with the SP-B domains was less than 50%.
Table 3 – Similarity of domain amino acid sequences of proteoglycans and Human surfactant proteins by results of paired alignment
|
|
Proteins |
PRG4 |
ACAN |
||||||
|
Proteins
|
Domains |
SMB1 |
SMB2 |
EGF-like |
Lectin C-type |
||||
|
% |
score |
% |
score |
% |
score |
% |
score |
||
|
SP-A
|
Collagen-like |
47 |
88 |
48 |
89 |
51 |
97 |
36 |
68 |
|
Lectin C-type |
39 |
73 |
39 |
73 |
41 |
77 |
52 |
98 |
|
|
SP-B |
Sasposin A1 |
60 |
113 |
61 |
114 |
62 |
117 |
38 |
72 |
|
Sasposin B1 |
57 |
108 |
58 |
109 |
57 |
107 |
39 |
73 |
|
|
Sasposin B2 |
59 |
110 |
62 |
116 |
62 |
116 |
38 |
71 |
|
|
Sasposin B3 |
60 |
113 |
60 |
113 |
62 |
117 |
41 |
77 |
|
|
SP-C |
BRICHOS |
45 |
85 |
47 |
88 |
48 |
90 |
32 |
60 |
|
SP-D |
Lectin C-type |
41 |
77 |
42 |
79 |
42 |
79 |
49 |
93 |
|
Collagen-like |
8 |
15 |
7 |
14 |
11 |
20 |
10 |
18 |
|
The domain similarity between SMB1 (PRG4), SMB2 (PRG4), EGF-like (ACAN) and BRICHOS (SP-C) was at the same level in the range from 45% to 48%. The minimum similarity value was recorded between the Lectin C-type of ACAN and the BRICHOS domain of SP-C.
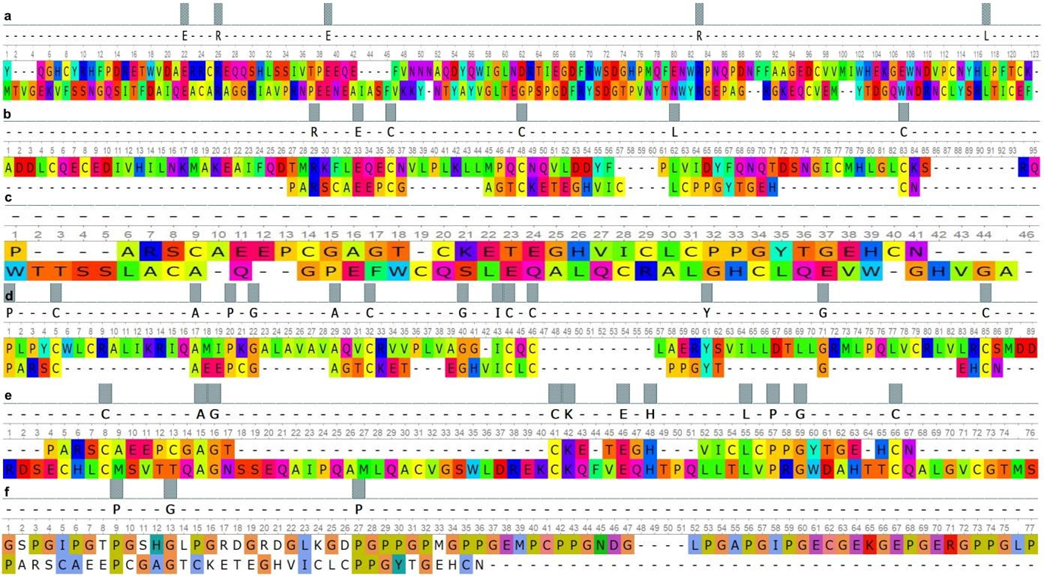
Fig. 1 – Paired alignment of ACAN domains and surfactant proteins:
a – ACAN_(Lectin_C-type) and SP-A_(Collagen-like); b – SP-B_(Sasposin_B1) and ACAN_(EGF-like); c – ACAN_(EGFlike) and SP-B_(Sasposin_A1); d – SP-B_(Sasposin_B2) and ACAN_(EGF-like); e – ACAN_(EGF-like) and SPB_(Sasposin_B3); d – SP-A_(Lectin_C-type) and ACAN_(EGF-like)
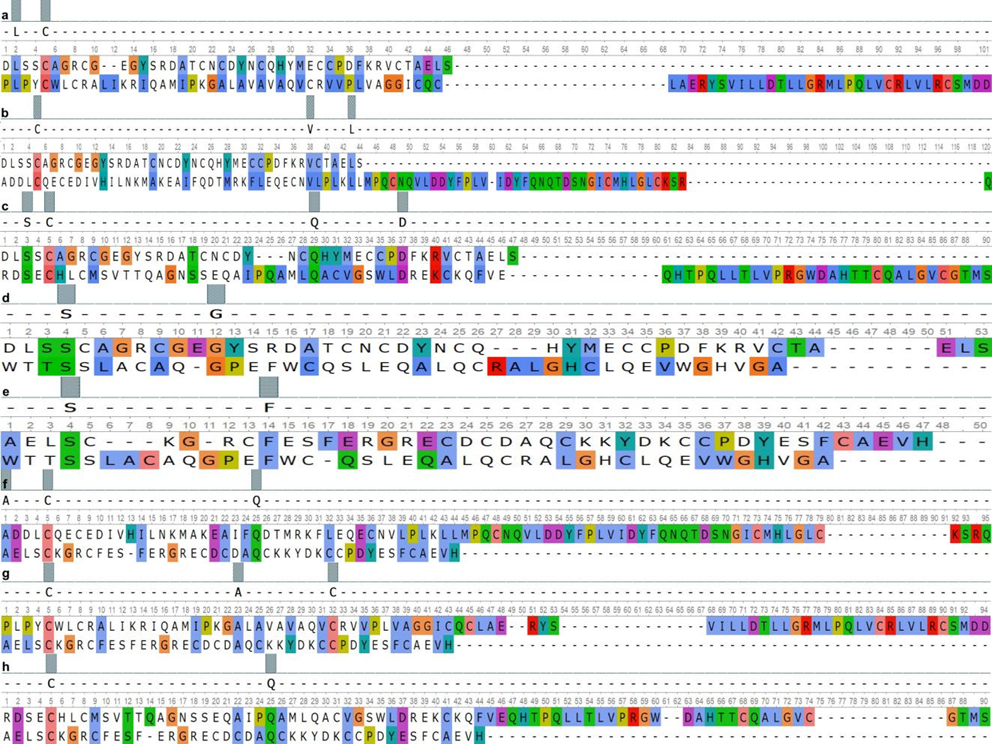
Fig. 2 – Paired alignment of PRG4 domains and surfactant proteins:
a – PRG4 (SMB1) and SP-B_(Sasposin_B2); b – PRG4 (SMB1) and SP-B_(Sasposin_B1); c – PRG4 (SMB1) and SPB_(Sasposin_B3); d – PRG4 (SMB1) and SP-B_(Sasposin_A1); e – PRG4 (SMB2) and SP-B_(Sasposin_A1); f – SPB_(Sasposin_B1) and PRG4 (SMB2); g – SP-B_(Sasposin_B2) and PRG4 (SMB2); h – SP-B_(Sasposin_B3) and PRG4 (SMB2) The homology between the SMB1 and SMB2 domains of PRG4 and Lectin C-type domain of SP-D was lower in comparison on the analogue alignment results for SP-A, but these differences data were not significant. At the same time, the Lectin C-type domain of the SP-D had higher similarity values with the same domain of the ACAN compared to SP-A. The lowest homology was found between the domains of the PRG4 and ACAN, and the Collagen-like domain of SP-D, which ranged from 7% to 11%.
According to the results of pair alignment, we found that the Lectin C-type domain, as well as the Collagen-like domain of SP-A and SP-D, had an insignificant structural similarity with the domains of PRG4 and ACAN, but it partially overlaps functionally [22]. According current researches, the first of these domains is a component of the extracellular matrix and is involved in intercellular adhesion [23], while the second one affects the decrease in surface tension [24]. The domains of the SPB protein had a high similarity with the studied domains of PRG4 and EGF-like domain of ACAN, which may indicate their common evolutionary origin. The BRICHOS domain is conservative, it provides the hydrophobic properties of the SP-C, although its functions are not fully understood, since it occurs in proteins belonging to different families [25]. At the same time, it is worth considering that the SP-C has the most similar functions to PRG4 [26].
3.3. The mucin domain in the structure of lubricin is PRG4
Multiple alignment of mucins (1-22) and PRG4 demonstrate a low degree of similarity between the proteins. Comparative analysis of the structure of mucins and PRG4 revealed PHA03247 domain (Table 4). This domain has been discovered in seven mucins as MUC-1, MUC-4, MUC-5B, MUC-6, MUC-16, MUC-19. Similar PRG4 domains were absent in the structure of other mucins.
Table 4 – Results of evaluation of structural homology between men and PRG4
|
Mucin (ID) |
Common domain with PRG4 |
Description |
Interval |
E-value |
|
MUC-1 (P15941) |
PHA03247
|
large tegument protein UL36; Provisional
|
247-797 |
7.84e-29 |
|
MUC-4 (Q99102) |
3301-3850 1330-1925 1040-1578 2876-3482 2124-2618 1735-2282 |
2.43e-18 1.40e-11 4.53e-06 5.09e-06 1.85e-05 1.15e-03 |
||
|
MUC-5B (Q9HC84) |
2594-2864 |
7.67e-03 |
||
|
MUC-6 (Q6W4X9) |
1215-1479 |
4.17e-07 |
||
|
MUC-16 (Q8WXI7) |
11503-11854 10751-11041 |
1.41e-06 6.09e-03 |
||
|
MUC-19 (Q7Z5P9) |
6651-6905 |
7.86e-03 |
The search for repeats that were experimentally found and had several variants [18] were found 32 times in five combinations (Fig. 3):
- |Е---Р---АРТТРК| = 18
- |К---Р---АРТТРК| = 6|
- |Е---Т---АРТТРК| = 3
- |G---Т---АРТТРК| = 1
- |K-S-APTTPK| = 4
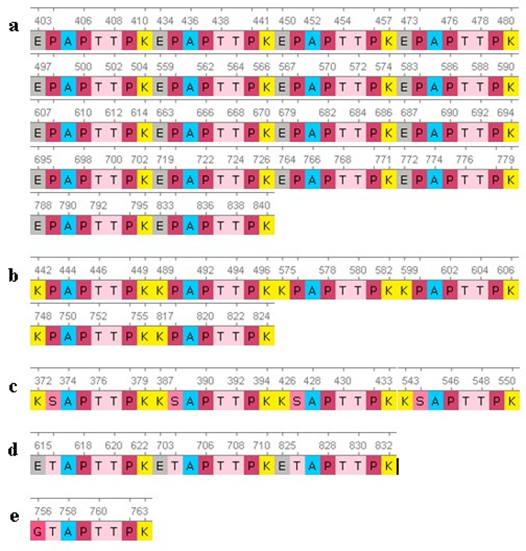
Fig. 3 – Sequences of repeats of the mucin domain in the protein PRG4:
a – «E---P---APTTPK»; b – «K---P---APTTPK»; Repeats «K---S---APTTPK»
Repeats «E---T---APTTPK». «G---T---APTTPK»
Repeat 1 was found in gaps along the entire length of the PRG4 molecule: [403-410], [434-441], [450-457], [473-480], [497504], [559-566], [567- 574], [583-590], [607-614], [663-670], [679-686], [687-694], [695-702], [719-726], [764-771], [772779], [788-795], [833-840].
Repeat 2 was identified at the following positions: [442-449], [489-496], [575-582], [599-606], [748-755], [817-824].
Repeat 3 was found only 3 times in the PRG4 molecule at the following locations: [615-622], [703-710], and [825-832].
Repeat 4 has only been encountered once in position [756-763].
Repeat 5 was identified at the following positions: [372-379,387-394,426-433,543-550].
From looking at the result analysis above, we can see that identified repeats didn't included in PRG4 domains. All of these repeats in the above positions also are not included the mucin (1-22).
According to results of multiple alignment of the full sequences of studied proteins, pair alignment of their domains the bilding of a phylogenetic tree became the next step of our study, to determine whether proteoglycans and surfactant proteins have a common evolutionary origin.
3.4. Phylogenetic analysis of proteoglycans and surfactant proteins and their human domains 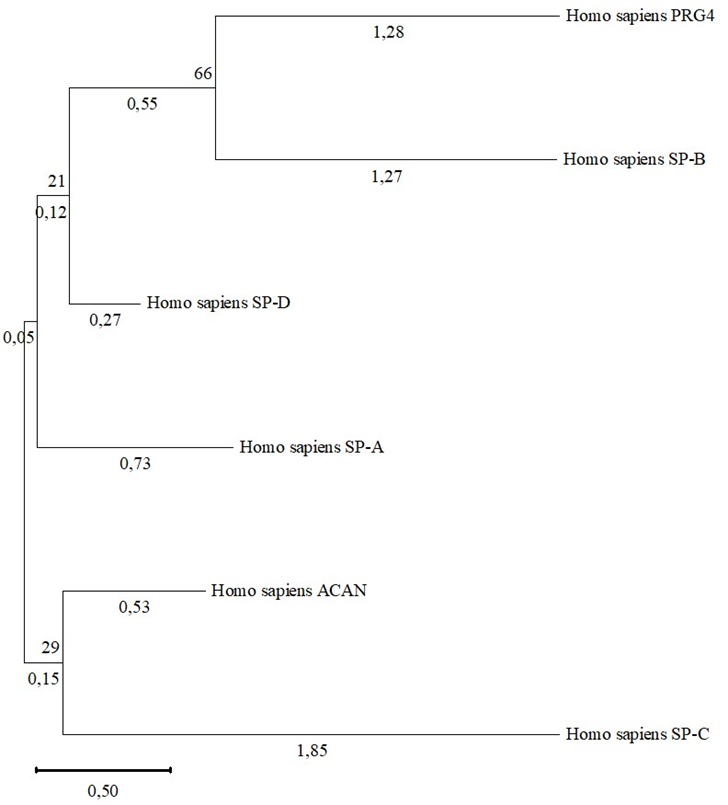
Fig. 4 – Phylogram of proteoglycans and human surfactant proteins
Note: shown next to the branches is the percentage of trees in which related taxa are grouped together
Other surfactant proteins both SP-D and SP-A are in a common clade with the PRG4 and SP-B, but they have very low bootstrap support, which in turn reduces the likelihood that this is a valid divergence.
The described results testify in favor of the fact that PRG4 and SP-B have a common origin, and this divergence occurred during the molecular evolution of these molecules, since they differ structurally in molecular length, but have similar functions. It is unclear the reason if the SP-C protein is not in a common clade with the PRG4 and SP-B, or it does not form a common clade with SP-B. However, low values of bootstrap support reduce the likelihood that this is a significant discrepancy.
Table 2 and Figure 5 demonstrate the resulting phylogram of Human PRG4, ACAN, SP-A, SP-B, SP-C, SP-D domains. As we can see, the clade with protein domains Sasposin B1, B2, B3 of SP-B has a high bootstrap support at 70, which is consistent with the fact that theese domains belong to the same family. A clade with a C-type lectin domain of SP-D and SP-A also has a high bootstrap support at 87. It is shown for PRG-4 domains and Sasposin A domain of the SP-B to have no evolutionary links with boottrap support 23, but this fact is not consistent with the phylogram of these proteins and the results of pair alignment.
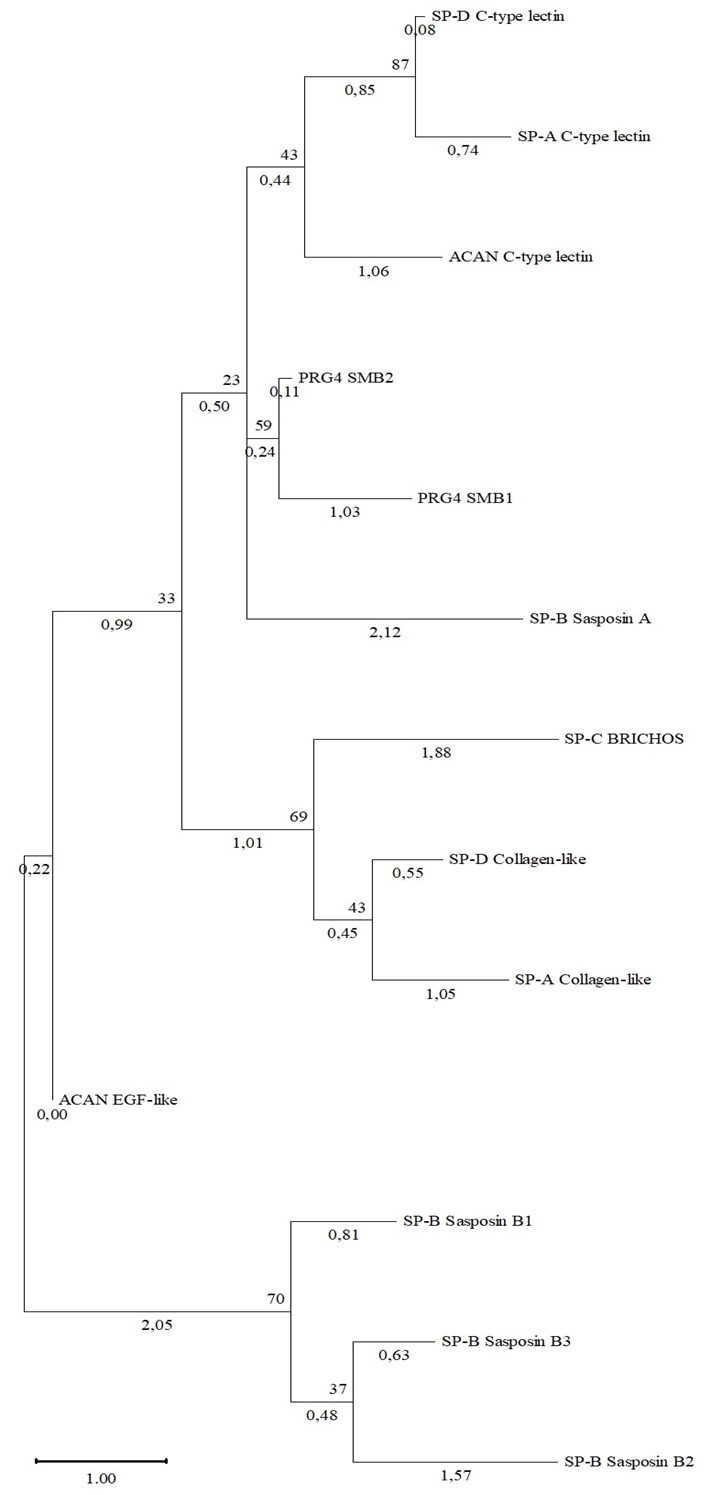
Fig. 5 – Phylogram of the domains of proteoglycans and human surfactant proteins
Note: shown next to the branches is the percentage of trees in which related taxa are grouped together
The strong differences can be explained by the different length of the amino acid sequences. These results may indicate on absence of structural homology of described proteins from the point of view of phylogeny,
4. Conclusion
The results revealed the presence of partial homology between proteoglycans and surfactant proteins. The high similarity between proteoglycans and surfactant proteins after multiple alignment was over 90% between Human and Papioanubis proteins, which is consistent with the results of protein molecular evolution. The smallest similarity was character for PRG4 of Human and Sus scrofa, it was less than 50%. On structural comparison of proteoglycans and surfactant proteins, the greatest similarity was found between the Sasposin A1, B1 B2, B3 domains of SP-B and SMB1, SMB2, EGF-like domains of PRG4 and ACAN. These indices were over 57%, which may indicate about a partial structural homology between the studied domains. The PRG4 of the studied animals is not characterized by the presence of mucin or mucin-like domains and repeats. Resultinf from phylogenetic analysis, the PRG4 и SP-B probably have common evolutionary origin.
Funding Финансирование
| The work was carried out with the support of a grant from the President of the Russian Federation for young scientists – candidates of science МК-199.2022.1.4. | Работа выполнена при поддержке гранта Президента Российской Федерации для молодых ученых – кандидатов наук МК-199.2022.1.4 |
Conflict of Interest Конфликт интересов
None declared. Не указан.
Список литературы
Saito T. The superficial zone of articular cartilage / T. Saito // Inflamm. Regen. — 2022. — Vol. 42, No. 1. — P. 14-19. — DOI: 10.1186/s41232-022-00202-0
Singh J. Surfactant protein disorders in childhood interstitial lung disease / J. Singh, A. Jaffe, A. Schultz [et al.] // Eur. J. Pediatr. — 2021. — Vol. 180, No 9. — P. 2711-2721. —DOI: 10.1007/s00431-021-04066-3
Grässel S. Recent advances in the treatment of osteoarthritis / S. Grässel, D. Muschter // F1000Research. — 2020. — No. 9 F1000. — P. Faculty Rev-325. — DOI: 10.12688/f1000research.22115.1
Ma L. Knee Osteoarthritis Therapy: Recent Advances in Intra-Articular Drug Delivery Systems / L. Ma, X. Zheng, R. Lin [et al.] // Drug design, development and therapy. — 2022. — Vol. 16. — P. 1311-1347. — DOI: 10.2147/DDDT.S357386
Novochadov V.V. Production technology and physicochemical properties of composition containing surfactant proteins / V.V. Novochadov, P.A. Krylov // European Journal of Molecular Biotechnology. — 2016. — No. 2(12). — P. 77-84. — DOI: 10.13187/ejmb.2016.12.77
Krylov P.A. Evaluation of the efficiency of lubricant based on pulmonary surfactant in experimental knee osteoarthritis in rats: analysis of 3D reconstructions / P.A. Krylov, A.S. Astakhov, E.N. Nesmeyanova [et al.] // Bulletin of Experimental Biology and Medicine. — 2020. — Vol. 168. — No. 3. — P. 371-374. — DOI: 10.1007/s10517-020-04711-1
Wu Y. Oxytocin prevents cartilage matrix destruction via regulating matrix metalloproteinases / Y. Wu, T. Wu, B. Xu [et al.] // Biochem. Biophys. Res. Commun. — 2017. — Vol. 486, No 3. — P. 601-606. — DOI: 10.1016/j.bbrc.2017.02.115
Fan X. Macro, micro, and molecular. changes of the osteochondral interface in osteoarthritis development / X. Fan, X. Wu, R. Crawford [et al.] // Front Cell Dev. Biol. — 2021. — Vol. 9. — e659654. — DOI:10.3389/fcell.2021.659654
Alekseeva L.I. Preparaty zamedlennogo dejstviya v lechenii osteoartroza [Delayed-acting drugs in the treatment of osteoarthritis] // Russian Medical Journal [Russkij Medicinskij ZHurnal]. — 2012. — Vol. 16. — № 7. — P. 389–391. [in Russian]
Boer C. G. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell / C. G. Boer, K. Hatzikotoulas, L. Southam — 2021. — Vol. 184, No 18. — P. 4784-4818. — e17. — DOI: 10.1016/j.cell.2021.07.038
Floros J. Differential regulation of human surfactant protein a genes, sftpa1 and sftpa2, and their corresponding variants / J. Floros, N. Tsotakos // Front Immunol. — 2021. — Vol. 12. — e766719. — DOI: 10.3389/fimmu.2021.766719
Hartjen N. Evaluation of surfactant proteins A, B, C, and D in articular cartilage, synovial membrane and synovial fluid of healthy as well as patients with osteoarthritis and rheumatoid arthritis / N. Hartjen, L. Bräuer, B. Reiß [et al.] // PLoS One. — 2018. — Vol. 13, No. 9. — e0203502. — DOI: 10.1371/journal.pone.0203502
Seime T. Proteoglycan 4 modulates osteogenic smooth muscle cell differentiation during vascular remodeling and intimal calcification / T. Seime, A.C. Akbulut, M.L. Liljeqvist [et al.] // Cells. — 2021. — Vol. 10, No 6. — e1276. —DOI: 10.3390/cells10061276
Menon N.G. Proteoglycan 4 (PRG4) expression and function in dry eye associated inflammation / N.G. Menon, R. Goyal, C. Lema // Exp. Eye Res. — 2021. — Vol. 208. — e108628. — DOI: 10.1016/j.exer.2021.108628
Akkiraju H. Role of chondrocytes in cartilage formation, progression of osteoarthritis and cartilage regeneration / H. Akkiraju, A. Nohe // J. Dev. Biol. — 2015. — Vol. 3, No. 4. — P. 177-192. — DOI: 10.3390/jdb3040177
Guagliardo R. Nanocarrier lipid composition modulates the impact of pulmonary surfactant protein B (SP-B) on cellular delivery of siRNA / R. Guagliardo, P. Merckx, A. Zamborlin [et al.] // Pharmaceutics. — 2019. — Vol. 11, No. 9. — e431. — DOI: 10.3390/pharmaceutics11090431
Sehlmeyer K. Alveolar dynamics and beyond – the importance of surfactant protein C and cholesterol in lung homeostasis and fibrosis / K. Sehlmeyer, J. Ruwisch, N. Roldan [et al.] // Front Physiol. — 2020. — Vol. 11. — e386. — DOI: 10.3389/fphys.2020.00386
Huang S. Cathepsin g degrades both glycosylated and unglycosylated regions of lubricin, a synovial mucin / S. Huang, K.A. Thomsson, C. Jin [et al.] // Sci. Rep. — 2020. — Vol.10, No. 1 — e4215. — DOI: 10.1038/s41598-020-61161-5
Dateki S. ACAN mutations as a cause of familial short stature / S. Dateki // Clin.Pediatr. Endocrinol. — 2017. — Vol. 26, No. 3. — P. 119-125. — DOI: 10.1297/cpe.26.119
King S.D. Recent progress on surfactant protein A: cellular function in lung and kidney disease development / S.D. King, S.Y. Chen // Am. J. Physiol. Cell Physiol. — 2020. — Vol. 319, No. 2. — P. C316-C320. — DOI: 10.1152/ajpcell.00195.2020
Johansson J. Synthetic surfactants with SP-B and SP-C analogues to enable worldwide treatment of neonatal respiratory distress syndrome and other lung diseases / J. Johansson, T. Curstedt // J. Intern. Med. — 2019. — Vol. 285, No. 2. — P. 165-186. — DOI: 10.1111/joim.12845
Sun Y. Altered autophagy in the mice with a deficiency of saposin A and saposin B / Y. Sun, G.A. Grabowski // Autophagy. — 2013. — Vol. 9, No. 7. — P. 1115-1116. — DOI: 10.4161/auto.24919
Ricard-Blum S. The collagen family / S. Ricard-Blum // Cold Spring HarbPerspect Biol. — 2011. — Vol. 3, No. 1. — a004978. — DOI: 10.1101/cshperspect.a004978
Ikegami M. The collagen-like region of surfactant protein A (SP-A) is required for correction of surfactant structural and functional defects in the SP-A null mouse / M. Ikegami, B.M. Elhalwagi, N. Palaniyar [et al.] // J. Biol. Chem. — 2001. — Vol. 276, No. 42. — P. 38542-38548. — DOI: 10.1074/jbc.M102054200
Hedlund J. Persson B. BRICHOS - a superfamily of multidomain proteins with diverse functions / J. Hedlund, J. Johansson, B. Persson [et al.] // BMC Res. Notes. — 2009. — Vol. 2. — e180. — DOI: 10.1186/1756-0500-2-180
Sáenz A. Folding and intramembraneous BRICHOS binding of the prosurfactant protein C transmembrane segment / A. Sáenz, J. Presto, P. Lara [et al.] // J. Biol. Chem. — 2015. — Vol. 290, No. 28. — P. 17628-17641. — DOI: 10.1074/jbc.M114.630343
UniProtKB [website]. — URL: https://www.uniprot.org/ (accessed: 20.11.2022)
NCBI Protein [website]. — URL: https://www.ncbi.nlm.nih.gov/protein (accessed: 20.11.2022)
EMBL-EBI [website]. — URL: https://www.ebi.ac.uk (accessed: 20.11.2022)
Ensembl [website]. — URL: https://www.ensembl.org/index.html (accessed: 20.11.2022)
NCBI [website]. — URL: https://www.ncbi.nlm.nih.gov/cdd/ (accessed: 20.11.2022)
