ОЦЕНКА СТРУКТУРНОЙ ГОМОЛОГИИ АБСЦИЗОВОЙ АЛЬДЕГИД-ОКСИДАЗЫ У ДРЕВЕСНЫХ РАСТЕНИЙ, ИСПОЛЬЗУЕМЫХ В АГРОЛЕСОМЕЛИОРАЦИИ НА ЮГЕ РОССИИ
ОЦЕНКА СТРУКТУРНОЙ ГОМОЛОГИИ АБСЦИЗОВОЙ АЛЬДЕГИД-ОКСИДАЗЫ У ДРЕВЕСНЫХ РАСТЕНИЙ, ИСПОЛЬЗУЕМЫХ В АГРОЛЕСОМЕЛИОРАЦИИ НА ЮГЕ РОССИИ
Аннотация
Деградация территорий и опустынивание приводят к сокращению разнообразия древесных растений. Поэтому важно увеличить видовое разнообразие в аридных районах путем внедрения новых видов. Для повышения эффективности ввода новых видов необходимо изучить генетические ресурсы древесных растений, устойчивых к неблагоприятным условиям среды. Целью работы было изучение фермента абсцизо-альдегид-оксидазы как потенциального генетического маркера у древесных растений, используемых в агролесомелиорации и при внедрении в аридные районы. Для получения результатов множественного выравнивания с помощью алгоритма ClustalW была использована программа UGENE, а филогенетическое дерево построено методом максимального правдоподобия с помощью программы MEGA 11. Аминокислотная последовательность фермента абсцизо-альдегид-оксидазы отличалась у Populus trichocarpa, который является наиболее засухоустойчивым видом среди исследованных деревьев. Филогенетический анализ этого фермента у Populus trichocarpa выявил эволюционные изменения и отличия в аминокислотной последовательности по сравнению с другими видами. Полученные результаты показали, что фермент абсцизо-альдегид-оксидаза может быть потенциальным генетическим маркером для определения засухоустойчивости растений. Фермент абсцизо-альдегид-оксидаза может быть использован в качестве одного из критериев отбора видов древесных растений для интродукции в засушливые климатические условия.
1. Introduction
Climate change leads to desertification and degradation of territories, which manifests itself in a decrease in the biodiversity of woody plants . In this regard, increasing the species diversity of arid territories through introduction is relevant . Increasing the efficiency of introduction involves searching for original and promising materials based on the study of the genetic resources of woody plants that are resistant to a complex of adverse environmental factors .
There are various criteria for the selection of promising woody plants, one of which is the physiological and biochemical parameters of the species . Currently, such a criterion as the molecular genetic criterion is actively used, which allows screening of resistant plant individuals at an early stage and studying the formation of adaptive reaction mechanisms at a fundamental level .
Abscisic-aldehyde oxidase (AAO) is an enzyme that plays an important role in plant life processes under various adverse environmental conditions. This enzyme is involved in the formation and metabolism of abscisic acid (ABA). ABA is a plant hormone that affects processes such as seed and fruit maturation, seed germination, stomatal closure, and adaptation to stress conditions , .
The role of AAO is to catalyze the oxidation of abscisic aldehyde to ABA. AAO is activated in response to stress, leading to an increase in ABA levels in plants. A high ABA level acts as a signal of external stress and activates a cascade of reactions aimed at protecting the plant from adverse environmental conditions. During periods of water stress, ABA levels increase, causing stomata to close to reduce water loss. Features of ABA synthesis and metabolism are also determined by the environmental conditions. For example, the activity of enzymes involved in ABA formation and conversion increases during drought or other stress. Therefore, AAO plays an important role in plant protection and adaptation to stress conditions , , .
The study of the role of AAO and ABA in plant metabolism is of great importance for understanding the mechanisms that ensure the survival and thriving of plants in arid conditions. Based on this, it is possible to select species for introduction into arid areas prone to desertification.
Thus, the aim of this study was to identify differences in the structural homology of AAO in native and introduced woody plants used in agroforestry in the southern territories of Russia.
2. Research methods and principles
The AAO enzyme was chosen as a potential gene marker whose expression may reflect changes in the state of drought-stressed plants. Woody species were selected from the forest seed bank organization program for protective afforestation in the southern regions of Russia , . Known drought-tolerant plants were taken into account. As a result, five plants with different numbers of AAO enzyme isoforms were found for further study: 2 isoforms of Malus sylvestris (wild apple), 3 isoforms of Populus trichocarpa (black cottonwood), 2 isoforms of Prunus mume (Japanese apricot), 1 isoform of Pyrus x bretschneideri (Chinese pear), and 2 isoforms of Quercus robur (common oak). Within the Populus genus, three species with the enzyme were found: Populus alba (white poplar), Populus euphratica (Euphrates poplar), and Populus trichocarpa (black cottonwood). To align the sequences, the amino acid sequences of the first isoforms of the selected plants were used:
1. Malus sylvestris: XP_050141529.1;
2. Populus alba A0A4V6A3J8;
3. Populus euphratica XP_011005929.1_1;
4. Populus trichocarpa: XP_024455533.2;
5. Prunus mume: XP_008230901.1;
6. Pyrus x bretschneideri: XP_048429325.1;
7. Quercus robur: XP_050284917.1.
Data on the structure of nucleotide and amino acid sequences of the studied genes of woody plants were obtained from the open-access databases UniProtKB , NCBI Protein , and NCBI Gene . Amino acid sequences were obtained based on the coding region of the gene. The search was conducted using synonymous constructs (Abscisic aldehyde oxidase[All Fields] AND (("Amorpha"[Organism] OR Amorpha[All Fields])]) OR ("Quercus"[Organism] OR Quercus[All Fields]) OR… ("Fraxinus"[Organism] OR Fraxinus[All Fields])) AND alive[prop]. Bioinformatic analysis was performed to identify differences between genes. It included multiple alignment of the amino acid sequences of the studied genes using the Ugene program (UNIPRO, Russia) and the ClustalW algorithm (BLOSUM62 matrix). The results of the alignment were evaluated by the percentage of identity, points (score), and the presence of common blocks (structural regions). Some sequences with the suffix “like” in the gene name were selected for alignment, as only such forms were present in the plant. This was because there were a limited number of responses to the query for the studied gene across the genera. Pseudogenes were not considered. A phylogenetic tree was constructed using the maximum likelihood method based on amino acid sequences, the JJT+G model, and bootstrap support in the MEGA11 program (MEGA, Japan).
3. Main results
As a result of analyzing the similarity of the enzyme in woody plants, we found potential differences that may affect the plant's ability to adapt to drought (see Table 1, Fig. 1, 2, 3).
Table 1 - Similarity of amino acid sequences of abscisic-aldehyde oxidase in woody plants according to the results of multiple alignment
| Species | Malus sylvestris | Populus trichocarpa | Prunus mume | Pyrus x bretschneideri | Quercus robur |
Species | Drought tolerance* | + | +++ | ++ | ++ | ++ |
Malus sylvestris | % | 100 | 69 | 83 | 97 | 75 |
score | 1409 | 972 | 1173 | 1363 | 1059 | |
Populus trichocarpa | % | 69 | 100 | 70 | 70 | 73 |
score | 972 | 1409 | 988 | 983 | 1034 | |
Prunus mume | % | 83 | 70 | 100 | 83 | 77 |
score | 1173 | 988 | 1409 | 1173 | 1078 | |
Pyrus x bretschneideri | % | 97 | 70 | 83 | 100 | 76 |
score | 1363 | 983 | 1173 | 1409 | 1068 | |
Quercus robur | % | 75 | 73 | 77 | 76 | 100 |
score | 1059 | 1034 | 1078 | 1068 | 1409 |
Note: «+++» - high, «++» - moderate, «+» - low drought tolerance
Populus trichocarpa is highly drought-resistant, while Malus sylvestris is poorly adapted to drought. The lowest percentage of similarity between the amino acid sequences of these trees is 69%. The similarity percentage between Populus trichocarpa and Prunus mume, Pyrus x bretschneideri, Quercus robur is approximately at the same level – 71%. The sequence of Malus sylvestris is most similar to Pyrus x bretschneideri. Prunus mume shows a high sequence similarity with both Malus sylvestris and Pyrus x bretschneideri. Since these trees are fruiting plants, their genetic sequences will be more similar to those of non-fruiting plants.

Figure 1 - Alignment of amino acid sequences of Malus sylvestris, Populus trichocarpa, Prunus mume, Pyrus x bretschneideri, and Quercus robur. Areas with a conservative level of more than 85% are colored dark gray
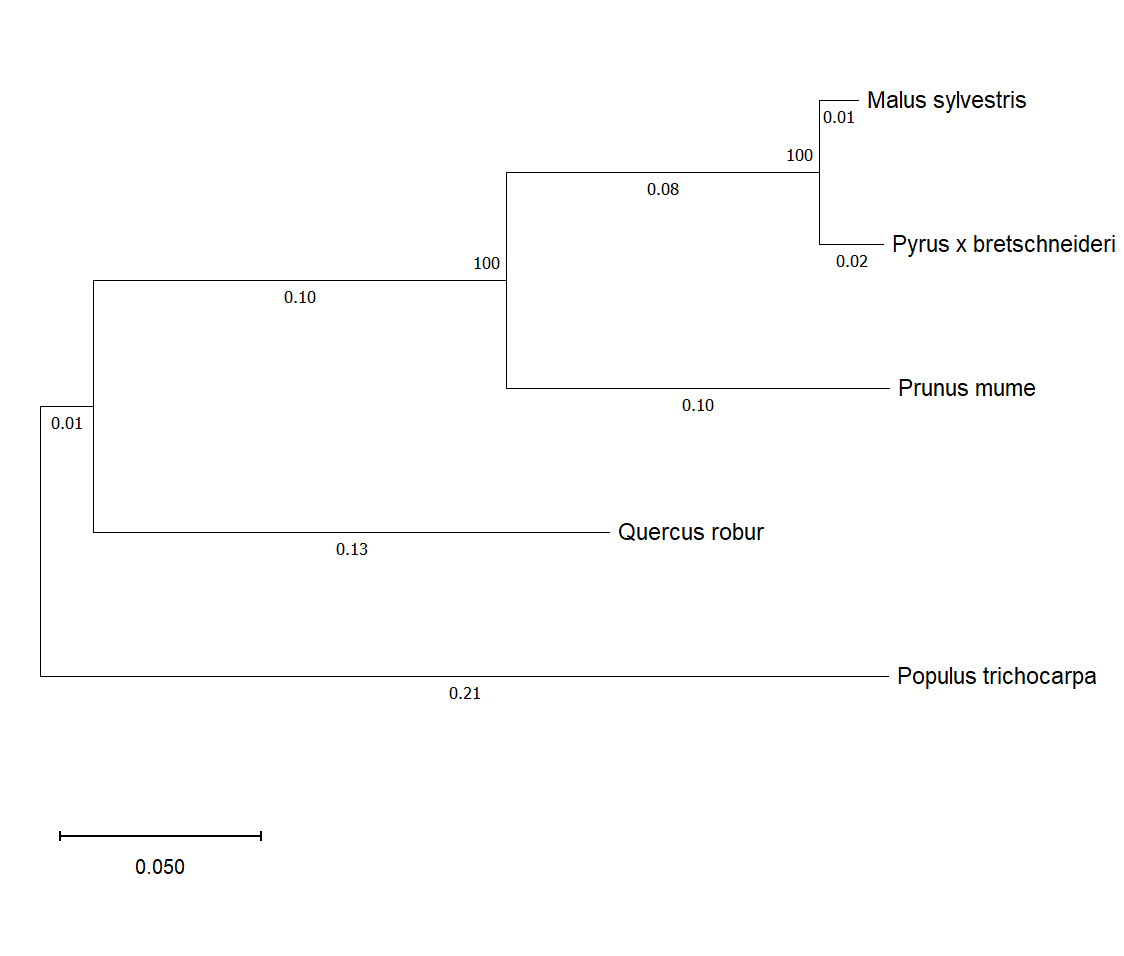
Figure 2 - Phylogram of the enzyme abscisic-aldehyde oxidase in woody plants
The evaluation of the structural homology of AAO in Malus sylvestris, Populus trichocarpa, Prunus mume, Pyrus x bretschneideri, and Quercus robur showed differences in several regions of the enzyme's amino acid sequence. This, in turn, may potentially influence the activity of the enzyme and the amount of its product involved in the formation of adaptive responses to drought , . Differences in the enzyme's structure were also found in Populus trichocarpa, which exhibits high drought resistance compared to other examined plants, potentially affecting its drought tolerance. Phylogenetic analysis demonstrated the distinctness of Populus trichocarpa from other tree species, which may indicate its genetic divergence from other plant species.
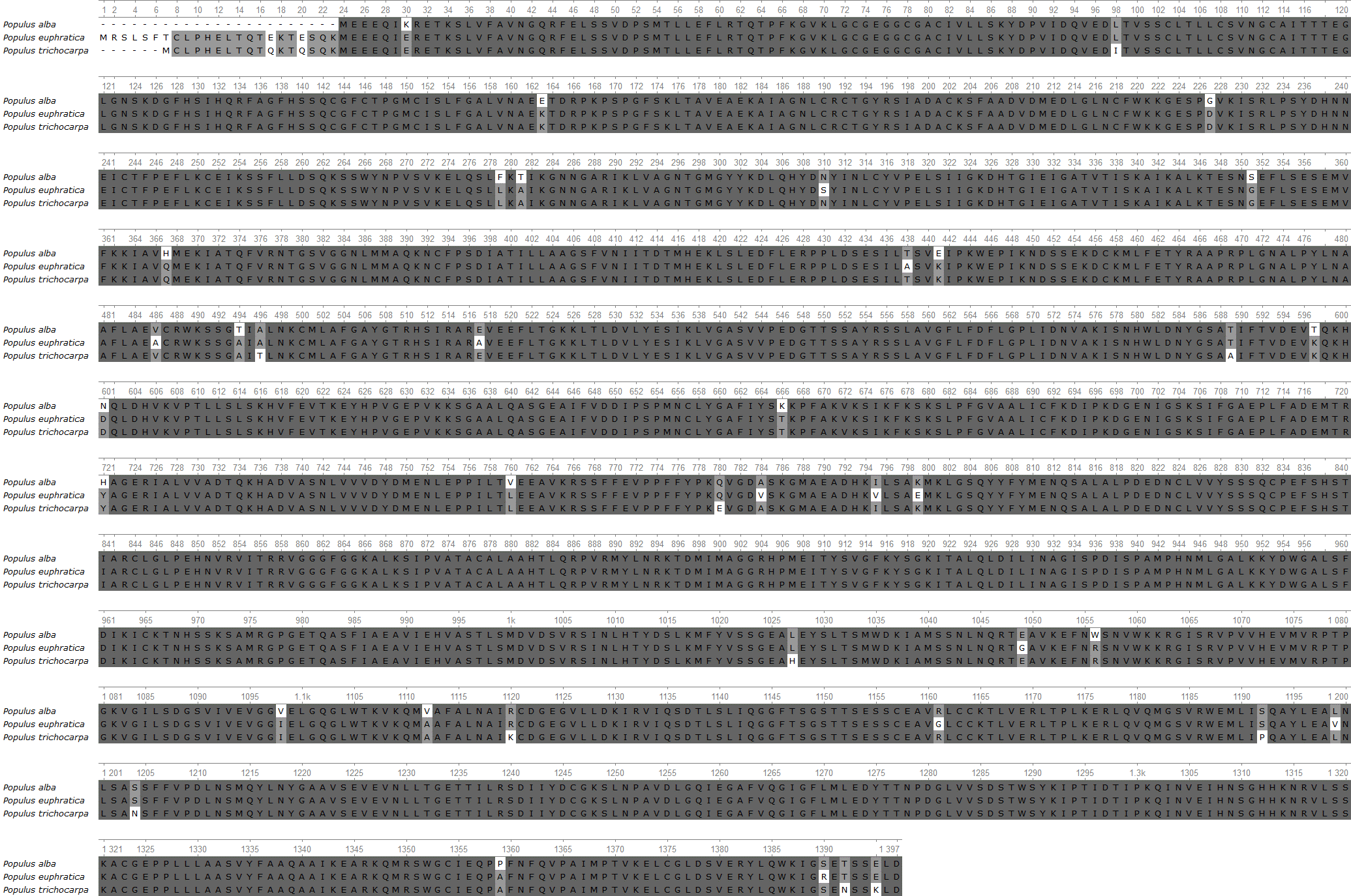
Figure 3 - Alignment of amino acid sequences of Populus alba, Populus euphratica, and Populus trichocarpa. Areas with a conservative level of more than 83% are colored dark gray
4. Discussion
To find the same distinctive positions in the genus Populus, multiple alignment of the enzyme was carried out. The analysis revealed that the sequences between poplar trees are more than 95% similar. The whole sequence is conservative; there are only single substitutions in positions. The domain structures also overlap . However, a region at the beginning of the sequences stands out: 1-23. Nevertheless, the genetic variability within the species can be further studied, as these variations may be associated with differences in ecological conditions or genetic diversity.
5. Conclusion
Populus trichocarpa is one of the most drought-tolerant plants among the studied species of woody plants. The amino acid sequence of the enzyme AAO in Populus trichocarpa differs partially from other studied woody plants. The presence of these unique differences may influence the functional properties of the enzyme. In addition, phylogenetic analysis based on this enzyme shows that Populus trichocarpa is relatively isolated from other studied plants. It can be hypothesized that Populus trichocarpa has evolved and developed a unique amino acid sequence of the AAO enzyme. The amino acid sequence similarity between Populus alba, Populus euphratica, and Populus trichocarpa is over 95%. It is unknown whether these differences have functional significance in the enzyme. Presumably, these changes may be related to environmental differences or are a result of genetic diversity among poplar species.
