Вернуться к статье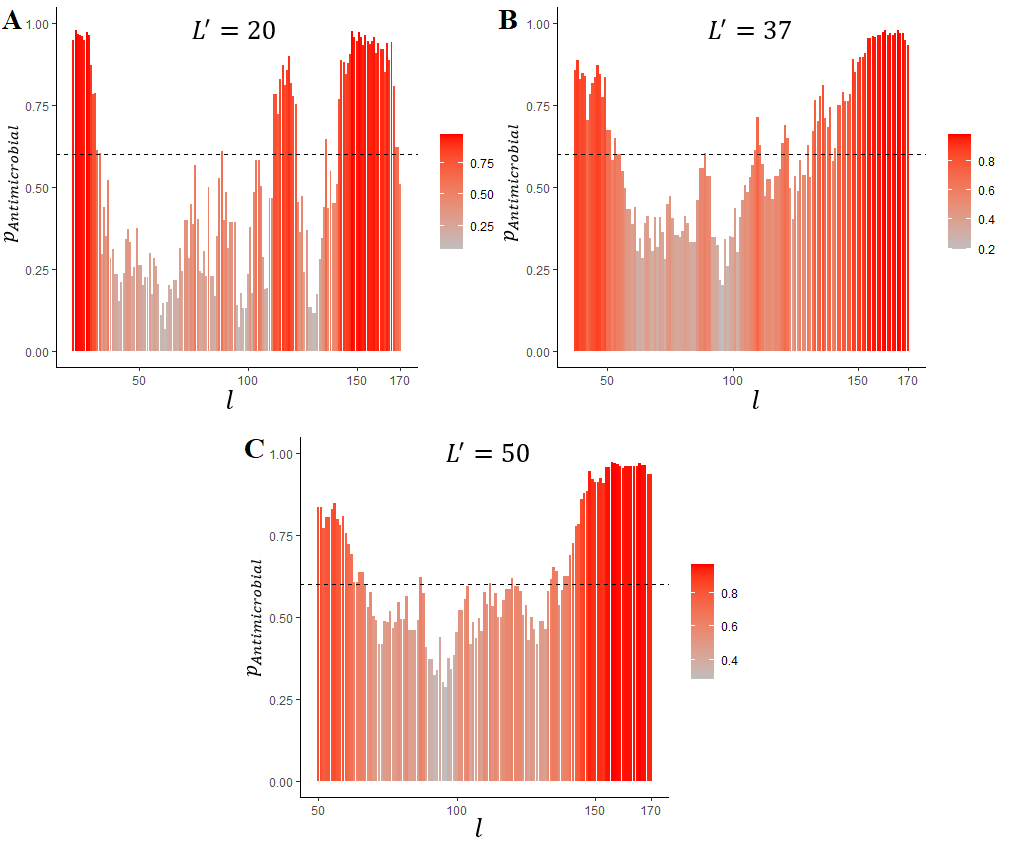
Машинное обучение с применением множественной логистической регрессии для предсказания антимикробных и гемолитических пептидов и их обнаружения в крупных белках
Figure 1 - Prediction of LL-37 localization in its precursor protein:
l is the rightmost a.a. in the sliding window, pAntimicrobial is probability of antimicrobial activity, L' is the sliding window length (A – 20, B – 37 and C – 50 amino acid a.a.)

the actual mature LL-37 length is 37 a.a. The dashed horizontal line represents the estimated optimal cut-off for AMPs prediction (0.6)
