ASSESSMENT OF THE HOMOLOGY OF THE AQUAPORIN IN REPRESENTATIVES OF SYMBIOTIC MYCORRHIZAL FUNGI
ASSESSMENT OF THE HOMOLOGY OF THE AQUAPORIN IN REPRESENTATIVES OF SYMBIOTIC MYCORRHIZAL FUNGI
Abstract
The presence of arbuscular mycorrhizae plays a crucial role in plant adaptation to drought. This process is associated with the aquaporins AQPF1 and AQPF2 in arbuscular mycorrhizal fungi (AMF). Therefore, this study evaluated the sequence homology of these two aquaporins (AQPF1 and AQPF2) in AMF using the BLAST program and the MUSCLE algorithm for multiple sequence alignment, followed by phylogenetic analysis with the MEGA12 software. The aim was to explore their potential mechanisms in regulating the water balance of the "fungus-plant" symbiosis under drought stress. The results demonstrated that AQPF1 exhibited significant sequence variability, establishing it as a promising candidate protein for investigating intraspecific variations related to drought resistance in arbuscular mycorrhizae.
1. Introduction
Globally, particularly in arid and semi-arid regions, water is the primary environmental factor limiting productivity in terrestrial ecosystems . Water stress inhibits photosynthesis, leading to reduced plant growth, diminished reproductive capacity, and mortality, consequently causing retrogressive succession of plant communities and species loss.
Arbuscular mycorrhizal (AM) fungi, belonging to the monophyletic phylum Glomeromycota, colonize over 80% of terrestrial plants and represent one of nature's most significant symbiotic fungi . Compared to non-mycorrhizal plants, mycorrhizal plants absorb water and nutrients not only directly through roots but also via the AM fungal pathway . AM fungi enhance host plant nutrient acquisition (particularly phosphorus), while regulating water balance and transport . Furthermore, AMF improve water-use efficiency (WUE) by modulating mechanisms that reduce oxidative damage under drought stress, offering a promising approach for sustainable agriculture . Studies indicate that AMF transport substantial water to host plants during drought .
Aquaporins constitute major pathways for transmembrane water movement and mediate nutrient-water exchange in mycorrhizal symbiosis . Research demonstrates that Glomus mosseae and Glomus intraradices downregulate aquaporin gene expression under drought. This downregulation enhances plant drought resistance by reducing membrane water permeability and facilitating cellular water retention .
The proteins AQPF1 and AQPF2 are two important aquaporins in AMF, and these proteins are related to the drought adaptation mechanisms initiated by symbiotic relationships . These proteins hold significant potential for elucidating drought resistance mechanisms in plant-fungal systems.
Therefore, this study aims to assess the amino acid sequence homology of aquaporins AQPF1 and AQPF2 across mycorrhizal fungi species to advance understanding of "fungus-plant" symbiotic dynamics under drought conditions.
2. Research methods and principles
The signaling proteins AQPF1 and AQPF2, identified across diverse arbuscular mycorrhizal fungi (AMF), were selected as research targets. Amino acid sequences for alignment construction were retrieved from the NCBI Protein database, and use the program BLAST to search for homologues of AQPF1 and AQPF2. The AQPF1 and AQPF2 protein sequence of Rhizophagus intraradices (with confirmed gene annotation and chromosomal localization) served as the initial query. Homologs of AQPF1 and AQPF2 were identified using the BLASTP program : Max Target Sequences 250, Matrix BLOSUM62, Gap Costs Existence 11, Extension 2. If annotated homologous sequences do not exist, then the closest unidentified homologous sequences are selected. Species with a similarity greater than 50% in the database were selected to construct the sequence alignment:
1) Rhizophagus intraradices (AFK93202.1);
2) Rhizophagus diaphanus MUCL43196 (RGB28923.1);
3) Rhizophagus irregularis (PKC58620.1);
4) Rhizophagus clarus (BAU71502.1);
5) Acaulospora morrowiae (CAG8589232.1);
6) Cetraspora pellucida (CAG8524555.1);
7) Diversispora epigaea (RHZ56822.1);
8) Diversispora eburnea (CAG8432930.1);
9) Dentiscutata erythropus (CAG8473267.1);
10) Dentiscutata heterogama (CAG8685703.1);
11) Funneliformis mosseae (CAG8578260.1);
12) Funneliformis caledonium (CAG8628874.1);
13) Funneliformis geosporum (CAI2164236.1);
14) Glomus cerebriforme (RIA94752.1);
15) Gigaspora margarita (KAF0497703.1);
16) Gigaspora rosea (RIB30586.1);
The species of the AQPF2 gene analyzed is exactly the same as those listed above. However, there are some minor differences in the protein names, which is due to the existence of known amino acid sequence information in the NCBI protein database
:1) Rhizophagus intraradices (AFK93203.1);
2) Rhizophagus diaphanus MUCL43196 (RGB43499.1);
3) Rhizophagus irregularis (XP_025170192.1);
4) Rhizophagus clarus (BAU71503.1);
5) Acaulospora morrowiae (CAG8574845.1);
6) Cetraspora pellucida (CAG8528323.1);
7) Diversispora epigaea (RHZ86381.1);
8) Diversispora eburnea (CAG8436721.1);
9) Dentiscutata erythropus (CAG8473267.1);
10) Dentiscutata heterogama (CAG8513861.1);
11) Funneliformis mosseae (CAG8590313.1);
12) Funneliformis caledonium (CAG8445309.1);
13) Funneliformis geosporum (CAI2179106.1);
14) Glomus cerebriforme (RIA98684.1);
15) Gigaspora margarita (CAG8854038.1);
16) Gigaspora rosea (RIB15405.1);
To identify differences between proteins, bioinformatic analysis was performed. It included multiple alignment of the amino acid sequences of the studied proteins using the MEGA 12 program (MEGA, Japan)
according to the MUSCLE algorithm with settings: penalty for gap opening -2.90, penalty for gap expansion 0, hydrophobicity multiplier 1.20. Alignment filtering of the sequences was completed manually with minimal changes: only the terminal unaligned regions were removed. Also, using MEGA 12, the most suitable model of amino acid substitutions was selected: LG + G for AQPF1 and AQPF2. Based on the amino acid sequences, a phylogenetic tree was constructed using the Neighbor-Joining method with bootstrap estimation -support at their default settings, in the MEGA 12 (MEGA, Japan).3. Main results
The multiple sequence alignment of AQPF2 revealed high conservation of this aquaporin, with a maximum pairwise genetic distance of 0.30167 observed between Acaulospora morrowiae and Glomus cerebriforme. No significant variable regions were detected at the N-terminus (Fig. 1). While the central region remained largely conserved across species, a 6-amino acid deletion was identified specifically in Diversispora epigaea and D. eburnea (Fig. 2). No other major indels were observed in this region.
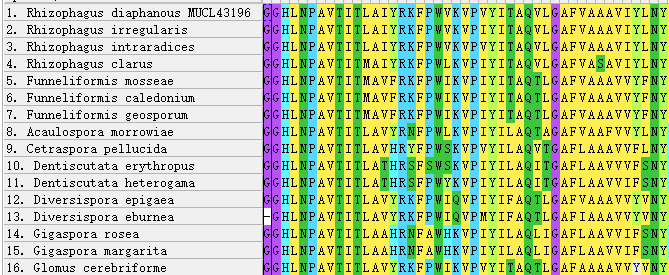
Figure 1 - N-terminus of AQPF2 alignment
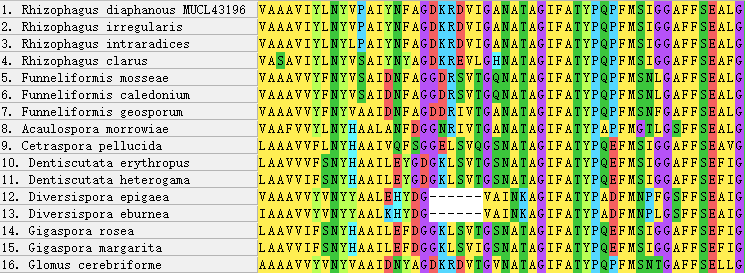
Figure 2 - Internal Deletion Region in the alignment of AQPF2 Protein
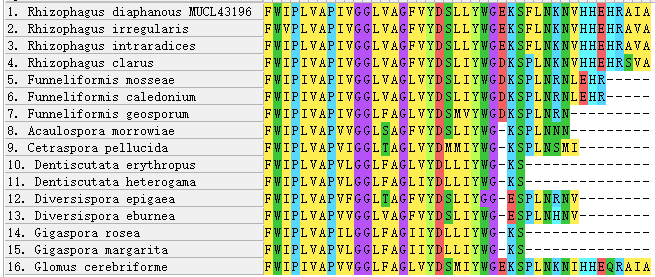
Figure 3 - C-terminus of AQPF2 alignment
The proteins that are most distant from the others are Diversispora epigaea and Diversispora eburnea from the Diversispora genus; their branch is longer than all other branches and is classified as a separate evolutionary branch.
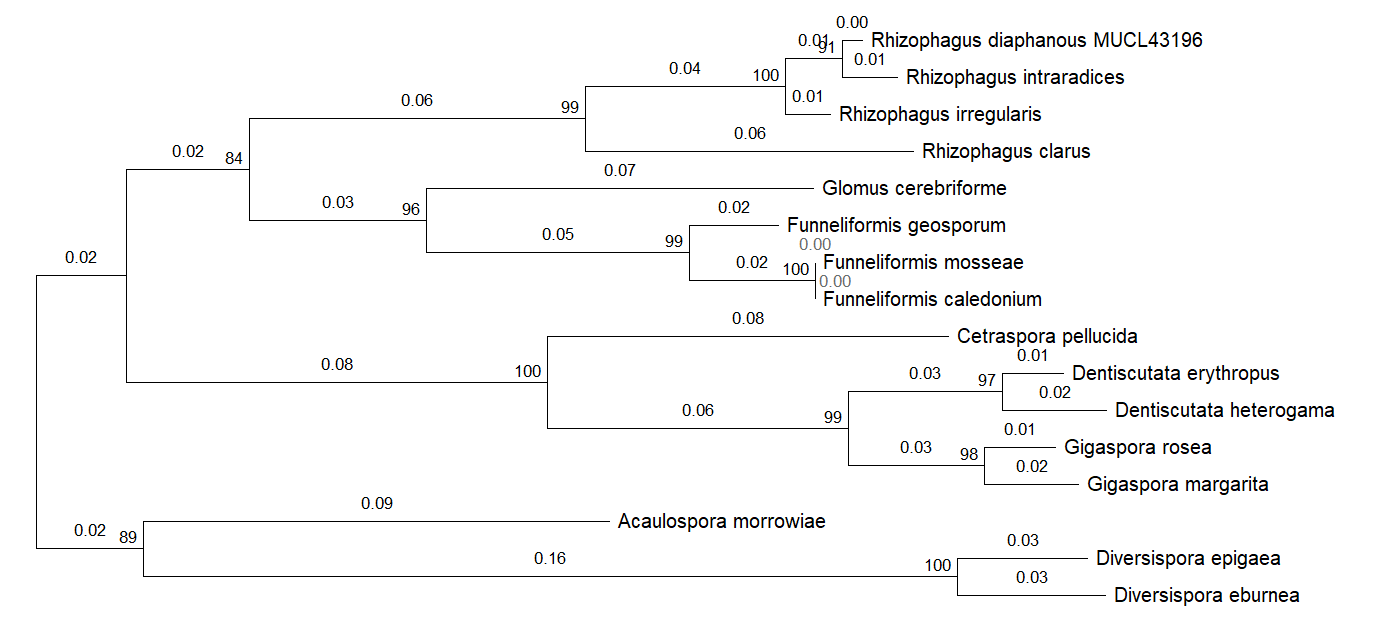
Figure 4 - AQPF2 proteins phylogenetic tree
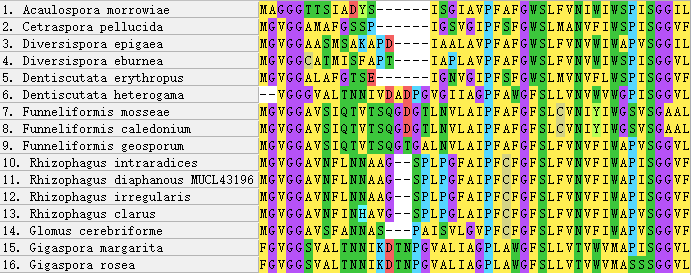
Figure 5 - N-terminus of AQPF1 alignment

Figure 6 - Insertions and variable regions in the alignment of AQPF1 Protein
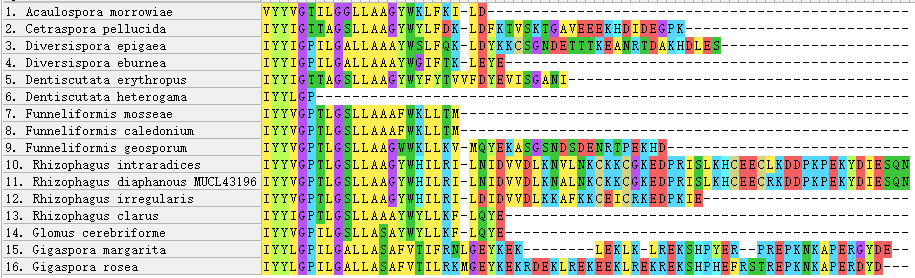
Figure 7 - C-terminus of AQPF1 alignment
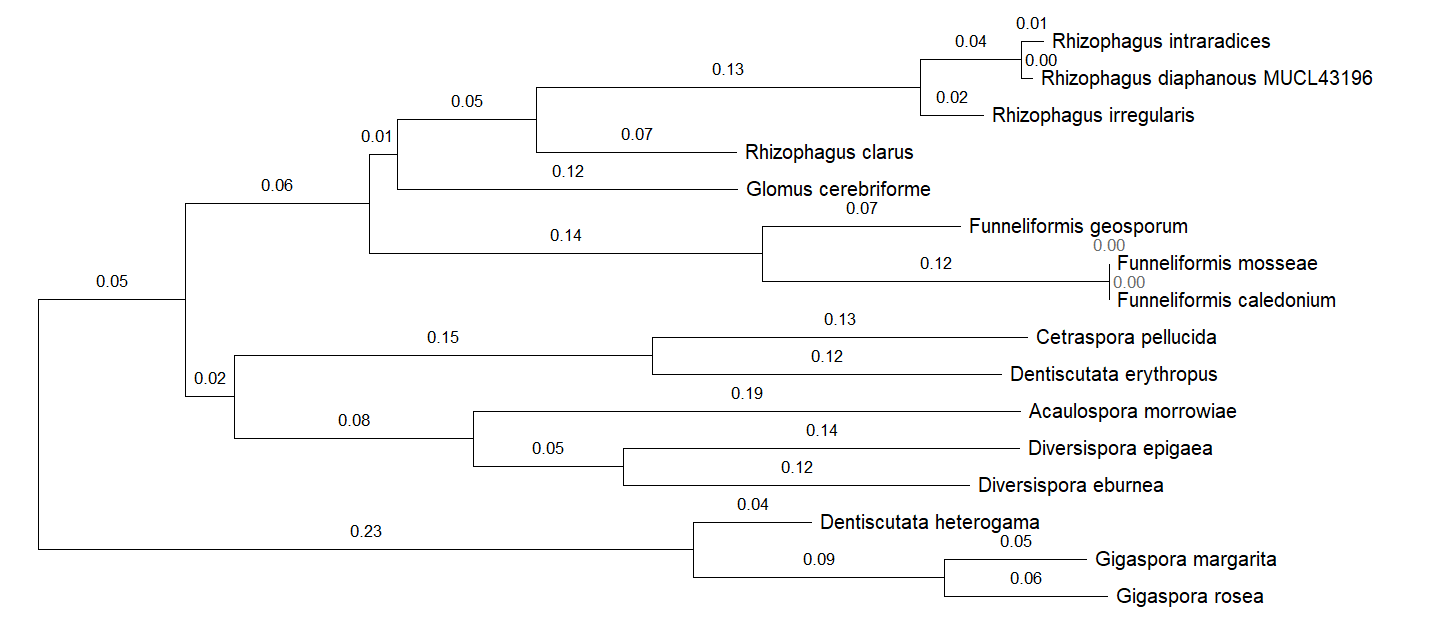
Figure 8 - AQPF1 proteins phylogenetic tree
4. Discussion
Compared to AQPF1, AQPF2 displays exceptional conservation. The presence of multiple paralogs in fungal genomes complicates phylogenetic reconstruction, and their functional divergence remains uncharacterized. Conversely, AQPF1 exhibits substantial sequence plasticity, with intra-Glomeromycota genetic distances typically exceeding 0.5. This structural variability positions AQPF1 as a priority target for elucidating AM fungal mechanisms in enhancing plant drought tolerance.
Notably, unlike typical proteins where variation localizes to terminal regions
, both terminal and core domains of AQPF1 demonstrate polymorphism. Although current data lack correlation between intra-species sequence variability and drought resistance phenotypes, future investigations could establish this linkage, potentially revealing aquaporin-mediated drought adaptation mechanisms.5. Conclusion
By comparing the two aquaporin proteins AQPF1 and AQPF2 and constructing a phylogenetic tree, the results revealed the conservation of the AQPF2 protein and the variability of the AQPF1 protein. AQPF1 has been identified as a promising protein for searching for intraspecific variations related to drought resistance.
